Long-Term Growth of Moss in Microfluidic Devices Enables Subcellular Studies in Development
- PMID: 27406170
- PMCID: PMC5074637
- DOI: 10.1104/pp.16.00879
Long-Term Growth of Moss in Microfluidic Devices Enables Subcellular Studies in Development
Abstract
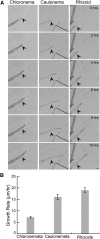
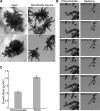
Key developmental processes that occur on the subcellular and cellular level or occur in occluded tissues are difficult to access, let alone image and analyze. Recently, culturing living samples within polydimethylsiloxane (PDMS) microfluidic devices has facilitated the study of hard-to-reach developmental events. Here, we show that an early diverging land plant, Physcomitrella patens, can be continuously cultured within PDMS microfluidic chambers. Because the PDMS chambers are bonded to a coverslip, it is possible to image P. patens development at high resolution over long time periods. Using PDMS chambers, we report that wild-type protonemal tissue grows at the same rate as previously reported for growth on solid medium. Using long-term imaging, we highlight key developmental events, demonstrate compatibility with high-resolution confocal microscopy, and obtain growth rates for a slow-growing mutant. By coupling the powerful genetic tools available to P. patens with long-term growth and imaging provided by PDMS microfluidic chambers, we demonstrate the capability to study cellular and subcellular developmental events in plants directly and in real time.
© 2016 American Society of Plant Biologists. All rights reserved.
Figures





References
-
- Dendukuri D, Panda P, Haghgooie R, Kim JM, Hatton TA, Doyle PS (2008) Modeling of oxygen-inhibited free radical photopolymerization in a PDMS microfluidic device. Macromolecules 41: 8547–8556
-
- Duffy DC, McDonald JC, Schueller OJ, Whitesides GM (1998) Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Anal Chem 70: 4974–4984 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

