A joint analysis of transcriptomic and metabolomic data uncovers enhanced enzyme-metabolite coupling in breast cancer
- PMID: 27406679
- PMCID: PMC4942812
- DOI: 10.1038/srep29662
A joint analysis of transcriptomic and metabolomic data uncovers enhanced enzyme-metabolite coupling in breast cancer
Abstract
Disrupted regulation of cellular processes is considered one of the hallmarks of cancer. We analyze metabolomic and transcriptomic profiles jointly collected from breast cancer and hepatocellular carcinoma patients to explore the associations between the expression of metabolic enzymes and the levels of the metabolites participating in the reactions they catalyze. Surprisingly, both breast cancer and hepatocellular tumors exhibit an increase in their gene-metabolites associations compared to noncancerous adjacent tissues. Following, we build predictors of metabolite levels from the expression of the enzyme genes catalyzing them. Applying these predictors to a large cohort of breast cancer samples we find that depleted levels of key cancer-related metabolites including glucose, glycine, serine and acetate are significantly associated with improved patient survival. Thus, we show that the levels of a wide range of metabolites in breast cancer can be successfully predicted from the transcriptome, going beyond the limited set of those measured.
Figures
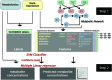
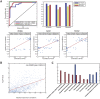
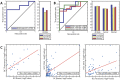
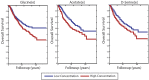
References
-
- Florian C. L., Preece N. E., Bhakoo K. K., Williams S. R. & Noble M. Characteristic metabolic profiles revealed by H-1 NMR spectroscopy for three types of human brain and nervous system tumours. NMR Biomed. 8, 253–264 (1995). - PubMed
-
- Tate a. R. et al.. Towards a method for automated classification of 1H MRS spectra from brain tumours. NMR Biomed. 11, 177–191 (1998). - PubMed
-
- McCarthy N. Metabolism: Unmasking an oncometabolite. Nat. Rev. Cancer 12, 229–229 (2012). - PubMed
-
- Sevin D. C., Kuehne A., Zamboni N. & Sauer U. Biological insights through nontargeted metabolomics. Curr. Opin. Biotechnol. 34, 1–8 (2015). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

