Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027
- PMID: 27406907
- PMCID: PMC5083130
- DOI: 10.1038/ja.2016.88
Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027
Abstract
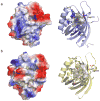
Comparative analysis of the enediyne biosynthetic gene clusters revealed sets of conserved genes serving as outstanding candidates for the enediyne core. Here we report the crystal structures of SgcJ and its homologue NCS-Orf16, together with gene inactivation and site-directed mutagenesis studies, to gain insight into enediyne core biosynthesis. Gene inactivation in vivo establishes that SgcJ is required for C-1027 production in Streptomyces globisporus. SgcJ and NCS-Orf16 share a common structure with the nuclear transport factor 2-like superfamily of proteins, featuring a putative substrate binding or catalytic active site. Site-directed mutagenesis of the conserved residues lining this site allowed us to propose that SgcJ and its homologues may play a catalytic role in transforming the linear polyene intermediate, along with other enediyne polyketide synthase-associated enzymes, into an enzyme-sequestered enediyne core intermediate. These findings will help formulate hypotheses and design experiments to ascertain the function of SgcJ and its homologues in nine-membered enediyne core biosynthesis.
Figures



Similar articles
-
A phosphopantetheinylating polyketide synthase producing a linear polyene to initiate enediyne antitumor antibiotic biosynthesis.Proc Natl Acad Sci U S A. 2008 Feb 5;105(5):1460-5. doi: 10.1073/pnas.0711625105. Epub 2008 Jan 25. Proc Natl Acad Sci U S A. 2008. PMID: 18223152 Free PMC article.
-
Biosynthesis of the enediyne antitumor antibiotic C-1027.Science. 2002 Aug 16;297(5584):1170-3. doi: 10.1126/science.1072110. Science. 2002. PMID: 12183628
-
Production of octaketide polyenes by the calicheamicin polyketide synthase CalE8: implications for the biosynthesis of enediyne core structures.J Am Chem Soc. 2009 Sep 9;131(35):12564-6. doi: 10.1021/ja904391r. J Am Chem Soc. 2009. PMID: 19689130 Free PMC article.
-
Iterative type I polyketide synthases involved in enediyne natural product biosynthesis.IUBMB Life. 2014 Sep;66(9):587-95. doi: 10.1002/iub.1316. Epub 2014 Oct 3. IUBMB Life. 2014. PMID: 25278375 Review.
-
Biosynthesis of enediyne antitumor antibiotics.Curr Top Med Chem. 2008;8(6):448-59. doi: 10.2174/156802608783955656. Curr Top Med Chem. 2008. PMID: 18397168 Free PMC article. Review.
Cited by
-
Comparative transcriptomic analysis reveals the significant pleiotropic regulatory effects of LmbU on lincomycin biosynthesis.Microb Cell Fact. 2020 Feb 12;19(1):30. doi: 10.1186/s12934-020-01298-0. Microb Cell Fact. 2020. PMID: 32050973 Free PMC article.
-
Conformational spread drives the evolution of the calcium-calmodulin protein kinase II.Sci Rep. 2022 May 19;12(1):8499. doi: 10.1038/s41598-022-12090-y. Sci Rep. 2022. PMID: 35589775 Free PMC article.
-
Structural characterization of three noncanonical NTF2-like superfamily proteins: implications for polyketide biosynthesis.Acta Crystallogr F Struct Biol Commun. 2020 Aug 1;76(Pt 8):372-383. doi: 10.1107/S2053230X20009814. Epub 2020 Jul 29. Acta Crystallogr F Struct Biol Commun. 2020. PMID: 32744249 Free PMC article.
-
Structure and Function of a Dual Reductase-Dehydratase Enzyme System Involved in p-Terphenyl Biosynthesis.ACS Chem Biol. 2021 Dec 17;16(12):2816-2824. doi: 10.1021/acschembio.1c00701. Epub 2021 Nov 11. ACS Chem Biol. 2021. PMID: 34763417 Free PMC article.
-
Mcc1229, an Stx2a-Amplifying Microcin, Is Produced In Vivo and Requires CirA for Activity.Infect Immun. 2022 Feb 17;90(2):e0058721. doi: 10.1128/IAI.00587-21. Epub 2021 Dec 6. Infect Immun. 2022. PMID: 34871041 Free PMC article.
References
-
- Liang ZX. Complexity and simplicity in the biosynthesis of enediyne natural products. Nat Prod Rep. 2010;27:499–528. - PubMed
-
- Edo K, et al. The structure of neocarzinostatin chromophore possessing a novel bicyclo [7,3,0]dodecadiyne system. Tetrahedron Lett. 1985;26:331–340.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

