A sodium channel inhibitor ISTX-I with a novel structure provides a new hint at the evolutionary link between two toxin folds
- PMID: 27407029
- PMCID: PMC4942781
- DOI: 10.1038/srep29691
A sodium channel inhibitor ISTX-I with a novel structure provides a new hint at the evolutionary link between two toxin folds
Abstract
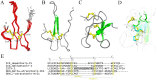
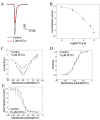
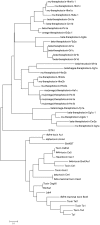
Members of arachnida, such as spiders and scorpions, commonly produce venom with specialized venom glands, paralyzing their prey with neurotoxins that specifically target ion channels. Two well-studied motifs, the disulfide-directed hairpin (DDH) and the inhibitor cystine knot motif (ICK), are both found in scorpion and spider toxins. As arachnids, ticks inject a neurotoxin-containing cocktail from their salivary glands into the host to acquire a blood meal, but peptide toxins acting on ion channels have not been observed in ticks. Here, a new neurotoxin (ISTX-I) that acts on sodium channels was identified from the hard tick Ixodes scapularis and characterized. ISTX-I exhibits a potent inhibitory function with an IC50 of 1.6 μM for sodium channel Nav1.7 but not other sodium channel subtypes. ISTX-I adopts a novel structural fold and is distinct from the canonical ICK motif. Analysis of the ISTX-I, DDH and ICK motifs reveals that the new ISTX-I motif might be an intermediate scaffold between DDH and ICK, and ISTX-I is a clue to the evolutionary link between the DDH and ICK motifs. These results provide a glimpse into the convergent evolution of neurotoxins from predatory and blood-sucking arthropods.
Figures


Similar articles
-
Holocyclotoxin-1, a cystine knot toxin from Ixodes holocyclus.Toxicon. 2014 Nov;90:308-17. doi: 10.1016/j.toxicon.2014.08.068. Epub 2014 Aug 27. Toxicon. 2014. PMID: 25172536
-
Solution structure of IsTX. A male scorpion toxin from Opisthacanthus madagascariensis (Ischnuridae).Eur J Biochem. 2004 Oct;271(19):3855-64. doi: 10.1111/j.1432-1033.2004.04322.x. Eur J Biochem. 2004. PMID: 15373831
-
Unique scorpion toxin with a putative ancestral fold provides insight into evolution of the inhibitor cystine knot motif.Proc Natl Acad Sci U S A. 2011 Jun 28;108(26):10478-83. doi: 10.1073/pnas.1103501108. Epub 2011 Jun 13. Proc Natl Acad Sci U S A. 2011. PMID: 21670253 Free PMC article.
-
Tarantulas: eight-legged pharmacists and combinatorial chemists.Toxicon. 2004 Apr;43(5):555-74. doi: 10.1016/j.toxicon.2004.02.007. Toxicon. 2004. PMID: 15066413 Review.
-
Insect-selective spider toxins targeting voltage-gated sodium channels.Toxicon. 2007 Mar 15;49(4):490-512. doi: 10.1016/j.toxicon.2006.11.027. Epub 2006 Dec 5. Toxicon. 2007. PMID: 17223149 Review.
Cited by
-
KNOTTIN: the database of inhibitor cystine knot scaffold after 10 years, toward a systematic structure modeling.Nucleic Acids Res. 2018 Jan 4;46(D1):D454-D458. doi: 10.1093/nar/gkx1084. Nucleic Acids Res. 2018. PMID: 29136213 Free PMC article.
-
Tick-Derived Peptide Blocks Potassium Channel TREK-1.Int J Mol Sci. 2024 Jul 31;25(15):8377. doi: 10.3390/ijms25158377. Int J Mol Sci. 2024. PMID: 39125945 Free PMC article.
-
Joannsin, a novel Kunitz-type FXa inhibitor from the venom of Prospirobolus joannsi.Thromb Haemost. 2017 Jun 2;117(6):1031-1039. doi: 10.1160/TH16-11-0829. Epub 2017 Mar 9. Thromb Haemost. 2017. PMID: 28276572 Free PMC article.
-
Tick Paralysis: Solving an Enigma.Vet Sci. 2018 May 14;5(2):53. doi: 10.3390/vetsci5020053. Vet Sci. 2018. PMID: 29757990 Free PMC article. Review.
References
-
- Platnick N. I. The world spider catalog, version 14.5. (American Museum of Natural History, 2014). Available at: http://research.amnh.org/entomology/spiders/catalog/index.html. 20/11/2014.
-
- Possani L. D., Becerril B., Delepierre M. & Tytgat J. Scorpion toxins specific for Na+-channels. Eur. J. Biochem. 264, 287–300 (1999). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

