DDK dependent regulation of TOP2A at centromeres revealed by a chemical genetics approach
- PMID: 27407105
- PMCID: PMC5062981
- DOI: 10.1093/nar/gkw626
DDK dependent regulation of TOP2A at centromeres revealed by a chemical genetics approach
Abstract
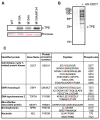
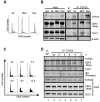
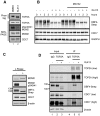
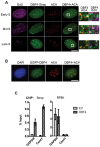
In eukaryotic cells the CDC7/DBF4 kinase, also known as DBF4-dependent kinase (DDK), is required for the firing of DNA replication origins. CDC7 is also involved in replication stress responses and its depletion sensitises cells to drugs that affect fork progression, including Topoisomerase 2 poisons. Although CDC7 is an important regulator of cell division, relatively few substrates and bona-fide CDC7 phosphorylation sites have been identified to date in human cells. In this study, we have generated an active recombinant CDC7/DBF4 kinase that can utilize bulky ATP analogues. By performing in vitro kinase assays using benzyl-thio-ATP, we have identified TOP2A as a primary CDC7 substrate in nuclear extracts, and serine 1213 and serine 1525 as in vitro phosphorylation sites. We show that CDC7/DBF4 and TOP2A interact in cells, that this interaction mainly occurs early in S-phase, and that it is compromised after treatment with CDC7 inhibitors. We further provide evidence that human DBF4 localises at centromeres, to which TOP2A is progressively recruited during S-phase. Importantly, we found that CDC7/DBF4 down-regulation, as well S1213A/S1525A TOP2A mutations can advance the timing of centromeric TOP2A recruitment in S-phase. Our results indicate that TOP2A is a novel DDK target and have important implications for centromere biology.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Localization of Cdc7 Protein Kinase During DNA Replication in Saccharomyces cerevisiae.G3 (Bethesda). 2017 Nov 6;7(11):3757-3774. doi: 10.1534/g3.117.300223. G3 (Bethesda). 2017. PMID: 28924058 Free PMC article.
-
Firing of Replication Origins Frees Dbf4-Cdc7 to Target Eco1 for Destruction.Curr Biol. 2017 Sep 25;27(18):2849-2855.e2. doi: 10.1016/j.cub.2017.07.070. Epub 2017 Sep 14. Curr Biol. 2017. PMID: 28918948 Free PMC article.
-
The Dbf4-Cdc7 kinase promotes Mcm2-7 ring opening to allow for single-stranded DNA extrusion and helicase assembly.J Biol Chem. 2015 Jan 9;290(2):1210-21. doi: 10.1074/jbc.M114.608232. Epub 2014 Dec 3. J Biol Chem. 2015. Retraction in: J Biol Chem. 2017 Jun 16;292(24):10320. doi: 10.1074/jbc.A114.608232. PMID: 25471369 Free PMC article. Retracted.
-
Regulation and roles of Cdc7 kinase under replication stress.Cell Cycle. 2014;13(12):1859-66. doi: 10.4161/cc.29251. Epub 2014 May 19. Cell Cycle. 2014. PMID: 24841992 Free PMC article. Review.
-
Novel insights into the roles of Cdc7 in response to replication stress.FEBS J. 2023 Jun;290(12):3076-3088. doi: 10.1111/febs.16456. Epub 2022 Apr 15. FEBS J. 2023. PMID: 35398961 Review.
Cited by
-
Mechanisms Governing DDK Regulation of the Initiation of DNA Replication.Genes (Basel). 2016 Dec 22;8(1):3. doi: 10.3390/genes8010003. Genes (Basel). 2016. PMID: 28025497 Free PMC article. Review.
-
A machine learning framework develops a DNA replication stress model for predicting clinical outcomes and therapeutic vulnerability in primary prostate cancer.J Transl Med. 2023 Jan 12;21(1):20. doi: 10.1186/s12967-023-03872-7. J Transl Med. 2023. PMID: 36635710 Free PMC article.
-
ZFP281-BRCA2 prevents R-loop accumulation during DNA replication.Nat Commun. 2022 Jun 17;13(1):3493. doi: 10.1038/s41467-022-31211-9. Nat Commun. 2022. PMID: 35715464 Free PMC article.
-
Propofol modulates the proliferation, invasion and migration of bladder cancer cells through the miR‑145‑5p/TOP2A axis.Mol Med Rep. 2021 Jun;23(6):439. doi: 10.3892/mmr.2021.12078. Epub 2021 Apr 13. Mol Med Rep. 2021. Retraction in: Mol Med Rep. 2024 Jun;29(6):94. doi: 10.3892/mmr.2024.13218. PMID: 33846791 Free PMC article. Retracted.
-
Exploration of the Role of the C-Terminal Domain of Human DNA Topoisomerase IIα in Catalytic Activity.ACS Omega. 2021 Sep 30;6(40):25892-25903. doi: 10.1021/acsomega.1c02083. eCollection 2021 Oct 12. ACS Omega. 2021. PMID: 34660952 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

