Novel small molecules potentiate premature termination codon readthrough by aminoglycosides
- PMID: 27407112
- PMCID: PMC5001621
- DOI: 10.1093/nar/gkw638
Novel small molecules potentiate premature termination codon readthrough by aminoglycosides
Abstract
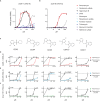
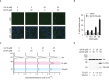
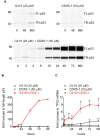
Nonsense mutations introduce premature termination codons and underlie 11% of genetic disease cases. High concentrations of aminoglycosides can restore gene function by eliciting premature termination codon readthrough but with low efficiency. Using a high-throughput screen, we identified compounds that potentiate readthrough by aminoglycosides at multiple nonsense alleles in yeast. Chemical optimization generated phthalimide derivative CDX5-1 with activity in human cells. Alone, CDX5-1 did not induce readthrough or increase TP53 mRNA levels in HDQ-P1 cancer cells with a homozygous TP53 nonsense mutation. However, in combination with aminoglycoside G418, it enhanced readthrough up to 180-fold over G418 alone. The combination also increased readthrough at all three nonsense codons in cancer cells with other TP53 nonsense mutations, as well as in cells from rare genetic disease patients with nonsense mutations in the CLN2, SMARCAL1 and DMD genes. These findings open up the possibility of treating patients across a spectrum of genetic diseases caused by nonsense mutations.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

