Multiple consecutive initiation of replication producing novel brush-like intermediates at the termini of linear viral dsDNA genomes with hairpin ends
- PMID: 27407114
- PMCID: PMC5062984
- DOI: 10.1093/nar/gkw636
Multiple consecutive initiation of replication producing novel brush-like intermediates at the termini of linear viral dsDNA genomes with hairpin ends
Abstract
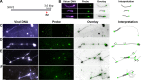
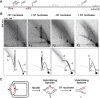
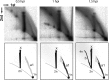
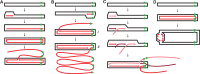
Linear dsDNA replicons with hairpin ends are found in the three domains of life, mainly associated with plasmids and viruses including the poxviruses, some phages and archaeal rudiviruses. However, their replication mechanism is not clearly understood. In this study, we find that the rudivirus SIRV2 undergoes multiple consecutive replication reinitiation events at the genomic termini. Using a strand-displacement replication strategy, the multiple reinitiation events from one parental template yield highly branched intermediates corresponding to about 30 genome units which generate exceptional 'brush-like' structures. Moreover, our data support the occurrence of an additional strand-coupled bidirectional replication from a circular dimeric intermediate. The multiple reinitiation process ensures rapid copying of the parental viral genome and will enable protein factors involved in viral genome replication to be specifically localised intracellularly, thereby helping the virus to avoid host defence mechanisms.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Sequences and replication of genomes of the archaeal rudiviruses SIRV1 and SIRV2: relationships to the archaeal lipothrixvirus SIFV and some eukaryal viruses.Virology. 2001 Dec 20;291(2):226-34. doi: 10.1006/viro.2001.1190. Virology. 2001. PMID: 11878892
-
Hairpin Transfer-Independent Parvovirus DNA Replication Produces Infectious Virus.J Virol. 2021 Sep 27;95(20):e0110821. doi: 10.1128/JVI.01108-21. Epub 2021 Aug 4. J Virol. 2021. PMID: 34346761 Free PMC article.
-
Stygiolobus rod-shaped virus and the interplay of crenarchaeal rudiviruses with the CRISPR antiviral system.J Bacteriol. 2008 Oct;190(20):6837-45. doi: 10.1128/JB.00795-08. Epub 2008 Aug 22. J Bacteriol. 2008. PMID: 18723627 Free PMC article.
-
Genomics and biology of Rudiviruses, a model for the study of virus-host interactions in Archaea.Biochem Soc Trans. 2013 Feb 1;41(1):443-50. doi: 10.1042/BST20120313. Biochem Soc Trans. 2013. PMID: 23356326 Free PMC article. Review.
-
Exceptionally diverse morphotypes and genomes of crenarchaeal hyperthermophilic viruses.Biochem Soc Trans. 2004 Apr;32(Pt 2):204-8. doi: 10.1042/bst0320204. Biochem Soc Trans. 2004. PMID: 15046572 Review.
Cited by
-
Viruses of archaea: Structural, functional, environmental and evolutionary genomics.Virus Res. 2018 Jan 15;244:181-193. doi: 10.1016/j.virusres.2017.11.025. Epub 2017 Nov 22. Virus Res. 2018. PMID: 29175107 Free PMC article. Review.
-
A filamentous archaeal virus is enveloped inside the cell and released through pyramidal portals.Proc Natl Acad Sci U S A. 2021 Aug 10;118(32):e2105540118. doi: 10.1073/pnas.2105540118. Proc Natl Acad Sci U S A. 2021. PMID: 34341107 Free PMC article.
-
Formation of a Viral Replication Focus in Sulfolobus Cells Infected by the Rudivirus Sulfolobus islandicus Rod-Shaped Virus 2.J Virol. 2017 Jun 9;91(13):e00486-17. doi: 10.1128/JVI.00486-17. Print 2017 Jul 1. J Virol. 2017. PMID: 28424282 Free PMC article.
-
A type III-B CRISPR-Cas effector complex mediating massive target DNA destruction.Nucleic Acids Res. 2017 Feb 28;45(4):1983-1993. doi: 10.1093/nar/gkw1274. Nucleic Acids Res. 2017. PMID: 27986854 Free PMC article.
-
The enigmatic archaeal virosphere.Nat Rev Microbiol. 2017 Nov 10;15(12):724-739. doi: 10.1038/nrmicro.2017.125. Nat Rev Microbiol. 2017. PMID: 29123227 Review.
References
-
- Weigel C., Seitz H. Bacteriophage replication modules. FEMS Microbiol. Rev. 2006;30:321–381. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

