DNA damage tolerance pathway involving DNA polymerase ι and the tumor suppressor p53 regulates DNA replication fork progression
- PMID: 27407148
- PMCID: PMC4968756
- DOI: 10.1073/pnas.1605828113
DNA damage tolerance pathway involving DNA polymerase ι and the tumor suppressor p53 regulates DNA replication fork progression
Abstract
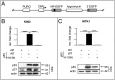
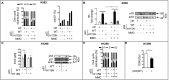
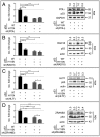
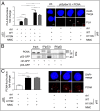
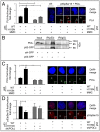
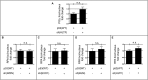
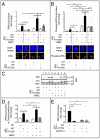
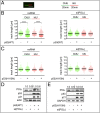
DNA damage tolerance facilitates the progression of replication forks that have encountered obstacles on the template strands. It involves either translesion DNA synthesis initiated by proliferating cell nuclear antigen monoubiquitination or less well-characterized fork reversal and template switch mechanisms. Herein, we characterize a novel tolerance pathway requiring the tumor suppressor p53, the translesion polymerase ι (POLι), the ubiquitin ligase Rad5-related helicase-like transcription factor (HLTF), and the SWI/SNF catalytic subunit (SNF2) translocase zinc finger ran-binding domain containing 3 (ZRANB3). This novel p53 activity is lost in the exonuclease-deficient but transcriptionally active p53(H115N) mutant. Wild-type p53, but not p53(H115N), associates with POLι in vivo. Strikingly, the concerted action of p53 and POLι decelerates nascent DNA elongation and promotes HLTF/ZRANB3-dependent recombination during unperturbed DNA replication. Particularly after cross-linker-induced replication stress, p53 and POLι also act together to promote meiotic recombination enzyme 11 (MRE11)-dependent accumulation of (phospho-)replication protein A (RPA)-coated ssDNA. These results implicate a direct role of p53 in the processing of replication forks encountering obstacles on the template strand. Our findings define an unprecedented function of p53 and POLι in the DNA damage response to endogenous or exogenous replication stress.
Keywords: DNA damage bypass; DNA polymerase idling; nascent DNA elongation; p53; polymerase ι.
Conflict of interest statement
Conflict of interest statement: L.W. is an inventor of a patent on a test system for determining genotoxicities, which is owned by L.W.
Figures



Similar articles
-
Multiple biochemical properties of the p53 molecule contribute to activation of polymerase iota-dependent DNA damage tolerance.Nucleic Acids Res. 2020 Dec 2;48(21):12188-12203. doi: 10.1093/nar/gkaa974. Nucleic Acids Res. 2020. PMID: 33166398 Free PMC article.
-
The levels of p53 govern the hierarchy of DNA damage tolerance pathway usage.Nucleic Acids Res. 2024 Apr 24;52(7):3740-3760. doi: 10.1093/nar/gkae061. Nucleic Acids Res. 2024. PMID: 38321962 Free PMC article.
-
p53 isoforms differentially impact on the POLι dependent DNA damage tolerance pathway.Cell Death Dis. 2021 Oct 13;12(10):941. doi: 10.1038/s41419-021-04224-3. Cell Death Dis. 2021. PMID: 34645785 Free PMC article.
-
Functions of SMARCAL1, ZRANB3, and HLTF in maintaining genome stability.Crit Rev Biochem Mol Biol. 2017 Dec;52(6):696-714. doi: 10.1080/10409238.2017.1380597. Epub 2017 Sep 28. Crit Rev Biochem Mol Biol. 2017. PMID: 28954549 Free PMC article. Review.
-
Post-replication repair: Rad5/HLTF regulation, activity on undamaged templates, and relationship to cancer.Crit Rev Biochem Mol Biol. 2019 Jun;54(3):301-332. doi: 10.1080/10409238.2019.1651817. Epub 2019 Aug 20. Crit Rev Biochem Mol Biol. 2019. PMID: 31429594 Review.
Cited by
-
Forging Ahead through Darkness: PCNA, Still the Principal Conductor at the Replication Fork.Mol Cell. 2017 Feb 2;65(3):380-392. doi: 10.1016/j.molcel.2016.12.020. Mol Cell. 2017. PMID: 28157503 Free PMC article. Review.
-
The Replication Stress Response on a Narrow Path Between Genomic Instability and Inflammation.Front Cell Dev Biol. 2021 Jun 25;9:702584. doi: 10.3389/fcell.2021.702584. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34249949 Free PMC article. Review.
-
Role of Translesion DNA Synthesis in the Metabolism of Replication-associated Nascent Strand Gaps.J Mol Biol. 2024 Jan 1;436(1):168275. doi: 10.1016/j.jmb.2023.168275. Epub 2023 Sep 13. J Mol Biol. 2024. PMID: 37714300 Free PMC article. Review.
-
Structural insights into the function of ZRANB3 in replication stress response.Nat Commun. 2017 Jun 16;8:15847. doi: 10.1038/ncomms15847. Nat Commun. 2017. PMID: 28621305 Free PMC article.
-
Nuclear PYHIN proteins target the host transcription factor Sp1 thereby restricting HIV-1 in human macrophages and CD4+ T cells.PLoS Pathog. 2020 Aug 6;16(8):e1008752. doi: 10.1371/journal.ppat.1008752. eCollection 2020 Aug. PLoS Pathog. 2020. PMID: 32760121 Free PMC article.
References
-
- Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358(6381):15–16. - PubMed
-
- Mummenbrauer T, et al. p53 protein exhibits 3′-to-5′ exonuclease activity. Cell. 1996;85(7):1089–1099. - PubMed
-
- Skalski V, Lin ZY, Choi BY, Brown KR. Substrate specificity of the p53-associated 3′-5′ exonuclease. Oncogene. 2000;19(29):3321–3329. - PubMed
-
- Bakhanashvili M. Exonucleolytic proofreading by p53 protein. Eur J Biochem. 2001;268(7):2047–2054. - PubMed
-
- Khare V, Eckert KA. The proofreading 3′-->5′ exonuclease activity of DNA polymerases: A kinetic barrier to translesion DNA synthesis. Mutat Res. 2002;510(1-2):45–54. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

