Thirty-five years of research into ribozymes and nucleic acid catalysis: where do we stand today?
- PMID: 27408700
- PMCID: PMC4926735
- DOI: 10.12688/f1000research.8601.1
Thirty-five years of research into ribozymes and nucleic acid catalysis: where do we stand today?
Abstract
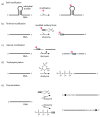
Since the discovery of the first catalytic RNA in 1981, the field of ribozyme research has developed from the discovery of catalytic RNA motifs in nature and the elucidation of their structures and catalytic mechanisms, into a field of engineering and design towards application in diagnostics, molecular biology and medicine. Owing to the development of powerful protocols for selection of nucleic acid catalysts with a desired functionality from random libraries, the spectrum of nucleic acid supported reactions has greatly enlarged, and importantly, ribozymes have been accompanied by DNAzymes. Current areas of research are the engineering of allosteric ribozymes for artificial regulation of gene expression, the design of ribozymes and DNAzymes for medicinal and environmental diagnostics, and the demonstration of RNA world relevant ribozyme activities. In addition, new catalytic motifs or novel genomic locations of known motifs continue to be discovered in all branches of life by the help of high-throughput bioinformatic approaches. Understanding the biological role of the catalytic RNA motifs widely distributed in diverse genetic contexts belongs to the big challenges of future RNA research.
Keywords: RNA; catalytic; ribozyme.
Conflict of interest statement
No competing interests were disclosed.
Figures




References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

