The Qatar genome: a population-specific tool for precision medicine in the Middle East
- PMID: 27408750
- PMCID: PMC4927697
- DOI: 10.1038/hgv.2016.16
The Qatar genome: a population-specific tool for precision medicine in the Middle East
Abstract
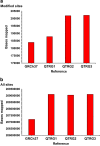
Reaching the full potential of precision medicine depends on the quality of personalized genome interpretation. In order to facilitate precision medicine in regions of the Middle East and North Africa (MENA), a population-specific genome for the indigenous Arab population of Qatar (QTRG) was constructed by incorporating allele frequency data from sequencing of 1,161 Qataris, representing 0.4% of the population. A total of 20.9 million single nucleotide polymorphisms (SNPs) and 3.1 million indels were observed in Qatar, including an average of 1.79% novel variants per individual genome. Replacement of the GRCh37 standard reference with QTRG in a best practices genome analysis workflow resulted in an average of 7* deeper coverage depth (an improvement of 23%) and 756,671 fewer variants on average, a reduction of 16% that is attributed to common Qatari alleles being present in QTRG. The benefit for using QTRG varies across ancestries, a factor that should be taken into consideration when selecting an appropriate reference for analysis.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

