The Genomic Sequence of the Oral Pathobiont Strain NI1060 Reveals Unique Strategies for Bacterial Competition and Pathogenicity
- PMID: 27409077
- PMCID: PMC4943601
- DOI: 10.1371/journal.pone.0158866
The Genomic Sequence of the Oral Pathobiont Strain NI1060 Reveals Unique Strategies for Bacterial Competition and Pathogenicity
Abstract
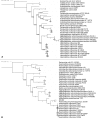
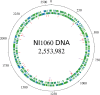
Strain NI1060 is an oral bacterium responsible for periodontitis in a murine ligature-induced disease model. To better understand its pathogenicity, we have determined the complete sequence of its 2,553,982 bp genome. Although closely related to Pasteurella pneumotropica, a pneumonia-associated rodent commensal based on its 16S rRNA, the NI1060 genomic content suggests that they are different species thriving on different energy sources via alternative metabolic pathways. Genomic and phylogenetic analyses showed that strain NI1060 is distinct from the genera currently described in the family Pasteurellaceae, and is likely to represent a novel species. In addition, we found putative virulence genes involved in lipooligosaccharide synthesis, adhesins and bacteriotoxic proteins. These genes are potentially important for host adaption and for the induction of dysbiosis through bacterial competition and pathogenicity. Importantly, strain NI1060 strongly stimulates Nod1, an innate immune receptor, but is defective in two peptidoglycan recycling genes due to a frameshift mutation. The in-depth analysis of its genome thus provides critical insights for the development of NI1060 as a prime model system for infectious disease.
Conflict of interest statement
Figures
References
-
- Marsh PD. Role of the Oral Microflora in Health. Microbial Ecology in Health and Disease. 2000. pp. 130–137. 10.1080/089106000750051800 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

