Induction of HOXA9 expression in three-dimensional organotypic culture of the Claudin-low breast cancer cells
- PMID: 27409175
- PMCID: PMC5239492
- DOI: 10.18632/oncotarget.10491
Induction of HOXA9 expression in three-dimensional organotypic culture of the Claudin-low breast cancer cells
Abstract
The gene expression signatures of the molecular intrinsic subtypes of breast cancer are regulated by epigenetic mechanisms such as methylation of CpG islands in gene promoters. Epigenetic codes can be regulated by the tumor microenvironment. The Claudin-low subtype is associated with triple-negative invasive ductal carcinomas in patients. Herein we explored epigenetic regulation of gene expression in the Claudin-low breast cancer cells by extracellular matrix (ECM), a key component of the tumor microenvironment. We modeled attachment to ECM using laminin rich ECM three-dimensional organotypic culture (lrECM 3D). In 2D and lrECM 3D cultures we examined expression of the homeobox (HOX) genes that epigenetically regulated in development and cancer. We demonstrated induction of the selected HOX genes in lrECM 3D culture of the Claudin-low breast cancer cells MDA-MB-231 and Hs578T. In particular activation of HOXA9 expression in lrECM 3D culture required binding of bromodomain containing 4 to the HOXA9 promoter and involved CpG hypomethylation. Our findings warrant further investigation of the ECM-regulated epigenetic coding of gene expression in the Claudin-low breast cancer.
Keywords: breast cancer; extracellular matrix; gene expression; homeobox gene; three-dimensional organotypic culture.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures
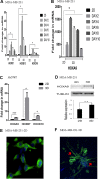
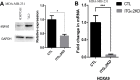
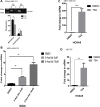
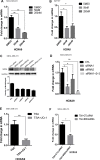
Similar articles
-
Induction of a novel isoform of the lncRNA HOTAIR in Claudin-low breast cancer cells attached to extracellular matrix.Mol Oncol. 2017 Dec;11(12):1698-1710. doi: 10.1002/1878-0261.12133. Epub 2017 Oct 30. Mol Oncol. 2017. PMID: 28846832 Free PMC article.
-
Epigenetic regulation of cancer stem cell genes in triple-negative breast cancer.Am J Pathol. 2012 Jul;181(1):257-67. doi: 10.1016/j.ajpath.2012.03.019. Epub 2012 May 21. Am J Pathol. 2012. PMID: 22626806
-
miR-7 and miR-218 epigenetically control tumor suppressor genes RASSF1A and Claudin-6 by targeting HoxB3 in breast cancer.Biochem Biophys Res Commun. 2012 Jul 20;424(1):28-33. doi: 10.1016/j.bbrc.2012.06.028. Epub 2012 Jun 14. Biochem Biophys Res Commun. 2012. PMID: 22705304
-
3D multicellular models to study the regulation and roles of acid-base transporters in breast cancer.Biochem Soc Trans. 2019 Dec 20;47(6):1689-1700. doi: 10.1042/BST20190131. Biochem Soc Trans. 2019. PMID: 31803922 Review.
-
[Is there a claudin-low phenotype of breast cancer?].Arkh Patol. 2022;84(1):45-49. doi: 10.17116/patol20228401145. Arkh Patol. 2022. PMID: 35166478 Review. Russian.
Cited by
-
The clinical significance of HOXA9 promoter hypermethylation in head and neck squamous cell carcinoma.J Clin Lab Anal. 2019 Jun;33(5):e22873. doi: 10.1002/jcla.22873. Epub 2019 Mar 6. J Clin Lab Anal. 2019. PMID: 30843252 Free PMC article.
-
Epigenetic Alterations in Triple-Negative Breast Cancer-The Critical Role of Extracellular Matrix.Cancers (Basel). 2021 Feb 9;13(4):713. doi: 10.3390/cancers13040713. Cancers (Basel). 2021. PMID: 33572395 Free PMC article. Review.
-
HOXC10 up-regulation promotes gastric cancer cell proliferation and metastasis through MAPK pathway.Chin J Cancer Res. 2017 Dec;29(6):572-580. doi: 10.21147/j.issn.1000-9604.2017.06.12. Chin J Cancer Res. 2017. PMID: 29353980 Free PMC article.
-
miR-133b suppresses metastasis by targeting HOXA9 in human colorectal cancer.Oncotarget. 2017 Jul 12;8(38):63935-63948. doi: 10.18632/oncotarget.19212. eCollection 2017 Sep 8. Oncotarget. 2017. PMID: 28969042 Free PMC article.
-
Roles of the HOX Proteins in Cancer Invasion and Metastasis.Cancers (Basel). 2020 Dec 22;13(1):10. doi: 10.3390/cancers13010010. Cancers (Basel). 2020. PMID: 33375038 Free PMC article. Review.
References
-
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
-
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

