A Quantitative Analysis of Complexity of Human Pathogen-Specific CD4 T Cell Responses in Healthy M. tuberculosis Infected South Africans
- PMID: 27409590
- PMCID: PMC4943605
- DOI: 10.1371/journal.ppat.1005760
A Quantitative Analysis of Complexity of Human Pathogen-Specific CD4 T Cell Responses in Healthy M. tuberculosis Infected South Africans
Abstract
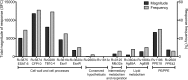
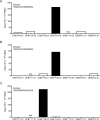
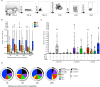
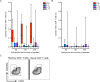
We performed a quantitative analysis of the HLA restriction, antigen and epitope specificity of human pathogen specific responses in healthy individuals infected with M. tuberculosis (Mtb), in a South African cohort as a test case. The results estimate the breadth of T cell responses for the first time in the context of an infection and human population setting. We determined the epitope repertoire of eleven representative Mtb antigens and a large panel of previously defined Mtb epitopes. We estimated that our analytic methods detected 50-75% of the total response in a cohort of 63 individuals. As expected, responses were highly heterogeneous, with responses to a total of 125 epitopes detected. The 66 top epitopes provided 80% coverage of the responses identified in our study. Using a panel of 48 HLA class II-transfected antigen-presenting cells, we determined HLA class II restrictions for 278 epitope/donor recognition events (36% of the total). The majority of epitopes were restricted by multiple HLA alleles, and 380 different epitope/HLA combinations comprised less than 30% of the estimated Mtb-specific response. Our results underline the complexity of human T cell responses at a population level. Efforts to capture and characterize this broad and highly HLA promiscuous Mtb-specific T cell epitope repertoire will require significant peptide multiplexing efforts. We show that a comprehensive "megapool" of Mtb peptides captured a large fraction of the Mtb-specific T cells and can be used to characterize this response.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Sallusto F, Geginat J, Lanzavecchia A (2004) Central Memory and Effector Memory T Cell Subsets: Function, Generation, and Maintenance. Annual Review of Immunology 22: 745–763. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

