Glial-cell-derived neuroregulators control type 3 innate lymphoid cells and gut defence
- PMID: 27409807
- PMCID: PMC4962913
- DOI: 10.1038/nature18644
Glial-cell-derived neuroregulators control type 3 innate lymphoid cells and gut defence
Abstract
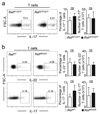
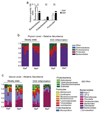
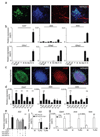
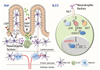
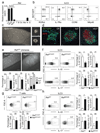
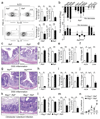
Group 3 innate lymphoid cells (ILC3) are major regulators of inflammation and infection at mucosal barriers. ILC3 development is thought to be programmed, but how ILC3 perceive, integrate and respond to local environmental signals remains unclear. Here we show that ILC3 in mice sense their environment and control gut defence as part of a glial–ILC3–epithelial cell unit orchestrated by neurotrophic factors. We found that enteric ILC3 express the neuroregulatory receptor RET. ILC3-autonomous Ret ablation led to decreased innate interleukin-22 (IL-22), impaired epithelial reactivity, dysbiosis and increased susceptibility to bowel inflammation and infection. Neurotrophic factors directly controlled innate Il22 downstream of the p38 MAPK/ERK-AKT cascade and STAT3 activation. Notably, ILC3 were adjacent to neurotrophic-factor-expressing glial cells that exhibited stellate-shaped projections into ILC3 aggregates. Glial cells sensed microenvironmental cues in a MYD88-dependent manner to control neurotrophic factors and innate IL-22. Accordingly, glial-intrinsic Myd88 deletion led to impaired production of ILC3-derived IL-22 and a pronounced propensity towards gut inflammation and infection. Our work sheds light on a novel multi-tissue defence unit, revealing that glial cells are central hubs of neuron and innate immune regulation by neurotrophic factor signals.
Conflict of interest statement
The authors declare no competing financial interests.
Figures








Comment in
-
Mucosal immunology: Glial cells support ILC3s and gut defence.Nat Rev Immunol. 2016 Jul 27;16(8):466-7. doi: 10.1038/nri.2016.86. Nat Rev Immunol. 2016. PMID: 27461150 No abstract available.
-
Immunology: A glia-ILC3 axis.Nat Rev Gastroenterol Hepatol. 2016 Sep;13(9):499. doi: 10.1038/nrgastro.2016.129. Epub 2016 Aug 3. Nat Rev Gastroenterol Hepatol. 2016. PMID: 27485790 No abstract available.
-
ILC3s and the Willow Tree of Voices.Immunity. 2016 Aug 16;45(2):238-9. doi: 10.1016/j.immuni.2016.08.004. Immunity. 2016. PMID: 27533011
-
A new edge to immune surveillance by the neural system.Cell Res. 2016 Nov;26(11):1178-1179. doi: 10.1038/cr.2016.107. Epub 2016 Sep 16. Cell Res. 2016. PMID: 27633062 Free PMC article.
-
OTULIN deficiency causes auto-inflammatory syndrome.Cell Res. 2016 Nov;26(11):1176-1177. doi: 10.1038/cr.2016.113. Epub 2016 Sep 30. Cell Res. 2016. PMID: 27686184 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

