Research Resource: A Reference Transcriptome for Constitutive Androstane Receptor and Pregnane X Receptor Xenobiotic Signaling
- PMID: 27409825
- PMCID: PMC4965847
- DOI: 10.1210/me.2016-1095
Research Resource: A Reference Transcriptome for Constitutive Androstane Receptor and Pregnane X Receptor Xenobiotic Signaling
Abstract
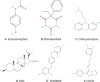
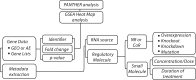
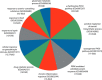
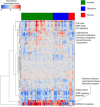
The pregnane X receptor (PXR) (PXR/NR1I3) and constitutive androstane receptor (CAR) (CAR/NR1I2) members of the nuclear receptor (NR) superfamily of ligand-regulated transcription factors are well-characterized mediators of xenobiotic and endocrine-disrupting chemical signaling. The Nuclear Receptor Signaling Atlas maintains a growing library of transcriptomic datasets involving perturbations of NR signaling pathways, many of which involve perturbations relevant to PXR and CAR xenobiotic signaling. Here, we generated a reference transcriptome based on the frequency of differential expression of genes across 159 experiments compiled from 22 datasets involving perturbations of CAR and PXR signaling pathways. In addition to the anticipated overrepresentation in the reference transcriptome of genes encoding components of the xenobiotic stress response, the ranking of genes involved in carbohydrate metabolism and gonadotropin action sheds mechanistic light on the suspected role of xenobiotics in metabolic syndrome and reproductive disorders. Gene Set Enrichment Analysis showed that although acetaminophen, chlorpromazine, and phenobarbital impacted many similar gene sets, differences in direction of regulation were evident in a variety of processes. Strikingly, gene sets representing genes linked to Parkinson's, Huntington's, and Alzheimer's diseases were enriched in all 3 transcriptomes. The reference xenobiotic transcriptome will be supplemented with additional future datasets to provide the community with a continually updated reference transcriptomic dataset for CAR- and PXR-mediated xenobiotic signaling. Our study demonstrates how aggregating and annotating transcriptomic datasets, and making them available for routine data mining, facilitates research into the mechanisms by which xenobiotics and endocrine-disrupting chemicals subvert conventional NR signaling modalities.
Figures
References
-
- Zhang J, Huang W, Chua SS, Wei P, Moore DD. Modulation of acetaminophen-induced hepatotoxicity by the xenobiotic receptor CAR. Science. 2002;298:422–424. - PubMed
-
- Guo GL, Moffit JS, Nicol CJ, et al. Enhanced acetaminophen toxicity by activation of the pregnane X receptor. Toxicol Sci. 2004;82:374–380. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

