Homeostatic Responses Regulate Selfish Mitochondrial Genome Dynamics in C. elegans
- PMID: 27411011
- PMCID: PMC5287496
- DOI: 10.1016/j.cmet.2016.06.008
Homeostatic Responses Regulate Selfish Mitochondrial Genome Dynamics in C. elegans
Abstract
Mutant mitochondrial genomes (mtDNA) can be viewed as selfish genetic elements that persist in a state of heteroplasmy despite having potentially deleterious metabolic consequences. We sought to study regulation of selfish mtDNA dynamics. We establish that the large 3.1-kb deletion-bearing mtDNA variant uaDf5 is a selfish genome in Caenorhabditis elegans. Next, we show that uaDf5 mutant mtDNA replicates in addition to, not at the expense of, wild-type mtDNA. These data suggest the existence of a homeostatic copy-number control that is exploited by uaDf5 to "hitchhike" to high frequency. We also observe activation of the mitochondrial unfolded protein response (UPR(mt)) in uaDf5 animals. Loss of UPR(mt) causes a decrease in uaDf5 frequency, whereas its constitutive activation increases uaDf5 levels. UPR(mt) activation protects uaDf5 from mitophagy. Taken together, we propose that mtDNA copy-number control and UPR(mt) represent two homeostatic response mechanisms that play important roles in regulating selfish mitochondrial genome dynamics.
Keywords: C. elegans; droplet digital PCR; evolution; heteroplasmy; mitochondrial unfolded protein response; mitophagy; mtDNA; mtDNA copy number control; selfish genetic element.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
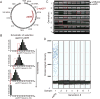
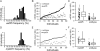


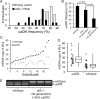


Comment in
-
Expanding Our Understanding of mtDNA Deletions.Cell Metab. 2016 Jul 12;24(1):3-4. doi: 10.1016/j.cmet.2016.06.024. Cell Metab. 2016. PMID: 27411002
References
-
- Al Rawi S, Louvet-Vallee S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–7. - PubMed
-
- Bernardi G. Lessons from a small, dispensable genome: the mitochondrial genome of yeast. Gene. 2005;354:189–200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

