Preventing Data Ambiguity in Infectious Diseases with Four-Dimensional and Personalized Evaluations
- PMID: 27411058
- PMCID: PMC4943638
- DOI: 10.1371/journal.pone.0159001
Preventing Data Ambiguity in Infectious Diseases with Four-Dimensional and Personalized Evaluations
Abstract
Background: Diagnostic errors can occur, in infectious diseases, when anti-microbial immune responses involve several temporal scales. When responses span from nanosecond to week and larger temporal scales, any pre-selected temporal scale is likely to miss some (faster or slower) responses. Hoping to prevent diagnostic errors, a pilot study was conducted to evaluate a four-dimensional (4D) method that captures the complexity and dynamics of infectious diseases.
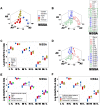
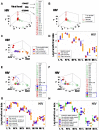
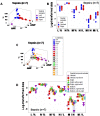
Methods: Leukocyte-microbial-temporal data were explored in canine and human (bacterial and/or viral) infections, with: (i) a non-structured approach, which measures leukocytes or microbes in isolation; and (ii) a structured method that assesses numerous combinations of interacting variables. Four alternatives of the structured method were tested: (i) a noise-reduction oriented version, which generates a single (one data point-wide) line of observations; (ii) a version that measures complex, three-dimensional (3D) data interactions; (iii) a non-numerical version that displays temporal data directionality (arrows that connect pairs of consecutive observations); and (iv) a full 4D (single line-, complexity-, directionality-based) version.
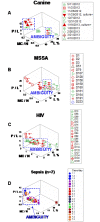
Results: In all studies, the non-structured approach revealed non-interpretable (ambiguous) data: observations numerically similar expressed different biological conditions, such as recovery and lack of recovery from infections. Ambiguity was also found when the data were structured as single lines. In contrast, two or more data subsets were distinguished and ambiguity was avoided when the data were structured as complex, 3D, single lines and, in addition, temporal data directionality was determined. The 4D method detected, even within one day, changes in immune profiles that occurred after antibiotics were prescribed.
Conclusions: Infectious disease data may be ambiguous. Four-dimensional methods may prevent ambiguity, providing earlier, in vivo, dynamic, complex, and personalized information that facilitates both diagnostics and selection or evaluation of anti-microbial therapies.
Conflict of interest statement
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

