Nosocomial transmission of Clostridium difficile ribotype 027 in a Chinese hospital, 2012-2014, traced by whole genome sequencing
- PMID: 27411304
- PMCID: PMC4942892
- DOI: 10.1186/s12864-016-2708-0
Nosocomial transmission of Clostridium difficile ribotype 027 in a Chinese hospital, 2012-2014, traced by whole genome sequencing
Abstract
Background: The rapid spread of Clostridium difficile NAP1/BI/027 (C. difficile 027) has become one of the leading threats of healthcare-associated infections worldwide. However, C. difficile 027 infections have been rarely reported in Asia, particularly in China.
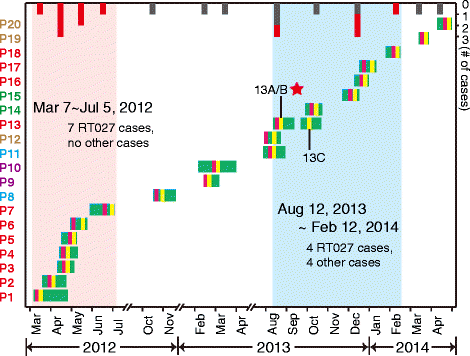
Results: In this study, we identified a rare C. difficile bloodstream infection (BSI) from three isolates of a patient during repeated hospital admission. This finding triggered a retrospective epidemiological study to scan all cases and strains emerged from this ward during the past three years. Using medical personnel interviews, medical record reviews and the genomic epidemiology, two outbreaks in 2012 and 2013-2014 were identified. Through using whole genome sequencing, we succeeded to trace the origin of the BSI strain. Surprisingly, we found the genome sequences were similar to C. difficile 027 strain R20291, indicating the occurrence of a rare C. difficile 027 strain in China. Integrated epidemiological investigation and whole genome sequencing of all strains, we constructed a nosocomial transmission map of these two C. difficile 027 outbreaks and traced the origin of the infection.
Conclusions: By genome sequencing, spatio-temporal analysis and field epidemiology investigation, we can estimate their complex transform network and reveal the possible modes of transmission in this ward. Based on their genetic diversity, we can assume that the toilets, bathroom, and janitor's equipment room may be contaminated area, which may be suggested to improve infection control measures in the following health care.
Keywords: Clostridium difficile; NAP1/BI/027; Nosocomial infection; Outbreak; Whole genome sequencing.
Figures



References
-
- Kuijper EJ, Barbut F, Brazier JS, et al. Update of Clostridium difficile infection due to PCR ribotype 027 in Europe, 2008. Euro Surveill 2008;13(31). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

