Quantitative Persulfide Site Identification (qPerS-SID) Reveals Protein Targets of H2S Releasing Donors in Mammalian Cells
- PMID: 27411966
- PMCID: PMC4944133
- DOI: 10.1038/srep29808
Quantitative Persulfide Site Identification (qPerS-SID) Reveals Protein Targets of H2S Releasing Donors in Mammalian Cells
Abstract
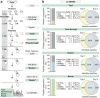
H2S is an important signalling molecule involved in diverse biological processes. It mediates the formation of cysteine persulfides (R-S-SH), which affect the activity of target proteins. Like thiols, persulfides show reactivity towards electrophiles and behave similarly to other cysteine modifications in a biotin switch assay. In this manuscript, we report on qPerS-SID a mass spectrometry-based method allowing the isolation of persulfide containing peptides in the mammalian proteome. With this method, we demonstrated that H2S donors differ in their efficacy to induce persulfides in HEK293 cells. Furthermore, data analysis revealed that persulfide formation affects all subcellular compartments and various cellular processes. Negatively charged amino acids appeared more frequently adjacent to cysteines forming persulfides. We confirmed our proteomic data using pyruvate kinase M2 as a model protein and showed that several cysteine residues are prone to persulfide formation finally leading to its inactivation. Taken together, the site-specific identification of persulfides on a proteome scale can help to identify target proteins involved in H2S signalling and enlightens the biology of H2S and its releasing agents.
Figures




References
-
- Bełtowski J. Hydrogen sulfide in pharmacology and medicine - An update. Pharmacol. Rep. 67, 647–658 (2015). - PubMed
-
- Li L. et al. Characterization of a novel, water-soluble hydrogen sulfide-releasing molecule (GYY4137): New insights into the biology of hydrogen sulfide. Circulation 117, 2351–2360 (2008). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

