The Many Roles of BAF (mSWI/SNF) and PBAF Complexes in Cancer
- PMID: 27413115
- PMCID: PMC4968166
- DOI: 10.1101/cshperspect.a026930
The Many Roles of BAF (mSWI/SNF) and PBAF Complexes in Cancer
Abstract
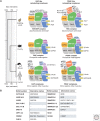
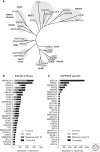
During the last decade, a host of epigenetic mechanisms were found to contribute to cancer and other human diseases. Several genomic studies have revealed that ∼20% of malignancies have alterations of the subunits of polymorphic BRG-/BRM-associated factor (BAF) and Polybromo-associated BAF (PBAF) complexes, making them among the most frequently mutated complexes in cancer. Recurrent mutations arise in genes encoding several BAF/PBAF subunits, including ARID1A, ARID2, PBRM1, SMARCA4, and SMARCB1 These subunits share some degree of conservation with subunits from related adenosine triphosphate (ATP)-dependent chromatin remodeling complexes in model organisms, in which a large body of work provides insight into their roles in cancer. Here, we review the roles of BAF- and PBAF-like complexes in these organisms, and relate these findings to recent discoveries in cancer epigenomics. We review several roles of BAF and PBAF complexes in cancer, including transcriptional regulation, DNA repair, and regulation of chromatin architecture and topology. More recent results highlight the need for new techniques to study these complexes.
Copyright © 2016 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures



References
-
- Aizawa H, Hu SC, Bobb K, Balakrishnan K, Ince G, Gurevich I, Cowan M, Ghosh A. 2004. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science 303: 197–202. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
