Pathogen-induced ERF68 regulates hypersensitive cell death in tomato
- PMID: 27415633
- PMCID: PMC6638261
- DOI: 10.1111/mpp.12460
Pathogen-induced ERF68 regulates hypersensitive cell death in tomato
Abstract
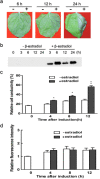
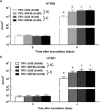
Ethylene response factors (ERFs) are a large plant-specific transcription factor family and play diverse important roles in various plant functions. However, most tomato ERFs have not been characterized. In this study, we showed that the expression of an uncharacterized member of the tomato ERF-IX subgroup, ERF68, was significantly induced by treatments with different bacterial pathogens, ethylene (ET) and salicylic acid (SA), but only slightly induced by bacterial mutants defective in the type III secretion system (T3SS) or non-host pathogens. The ERF68-green fluorescent protein (ERF68-GFP) fusion protein was localized in the nucleus. Transactivation and electrophoretic mobility shift assays (EMSAs) further showed that ERF68 was a functional transcriptional activator and was bound to the GCC-box. Moreover, transient overexpression of ERF68 led to spontaneous lesions in tomato and tobacco leaves and enhanced the expression of genes involved in ET, SA, jasmonic acid (JA) and hypersensitive response (HR) pathways, whereas silencing of ERF68 increased tomato susceptibility to two incompatible Xanthomonas spp. These results reveal the involvement of ERF68 in the effector-triggered immunity (ETI) pathway. To identify ERF68 target genes, chromatin immunoprecipitation combined with high-throughput sequencing (ChIP-seq) was performed. Amongst the confirmed target genes, a few genes involved in cell death or disease defence were differentially regulated by ERF68. Our study demonstrates the function of ERF68 in the positive regulation of hypersensitive cell death and disease defence by modulation of multiple signalling pathways, and provides important new information on the complex regulatory function of ERFs.
Keywords: ERF; cell death; defence; disease; tomato.
© 2016 BSPP AND JOHN WILEY & SONS LTD.
Figures




Similar articles
-
SlBIR3 Negatively Regulates PAMP Responses and Cell Death in Tomato.Int J Mol Sci. 2017 Sep 13;18(9):1966. doi: 10.3390/ijms18091966. Int J Mol Sci. 2017. PMID: 28902164 Free PMC article.
-
Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum.Plant Mol Biol. 2004 Aug;55(6):825-34. doi: 10.1007/s11103-004-2140-8. Plant Mol Biol. 2004. PMID: 15604719
-
Functional analysis and binding affinity of tomato ethylene response factors provide insight on the molecular bases of plant differential responses to ethylene.BMC Plant Biol. 2012 Oct 11;12:190. doi: 10.1186/1471-2229-12-190. BMC Plant Biol. 2012. PMID: 23057995 Free PMC article.
-
Durability of resistance in tomato and pepper to xanthomonads causing bacterial spot.Annu Rev Phytopathol. 2009;47:265-84. doi: 10.1146/annurev-phyto-080508-081752. Annu Rev Phytopathol. 2009. PMID: 19400644 Review.
-
Signal recognition and transduction involved in plant disease resistance.Essays Biochem. 1997;32:87-99. Essays Biochem. 1997. PMID: 9493013 Review.
Cited by
-
Comparison of Tomato Transcriptomic Profiles Reveals Overlapping Patterns in Abiotic and Biotic Stress Responses.Int J Mol Sci. 2023 Feb 17;24(4):4061. doi: 10.3390/ijms24044061. Int J Mol Sci. 2023. PMID: 36835470 Free PMC article.
-
A virulent milRNA of Fusarium oxysporum f. sp. cubense impairs plant resistance by targeting banana AP2 transcription factor coding gene MaPTI6L.Hortic Res. 2024 Dec 28;12(4):uhae361. doi: 10.1093/hr/uhae361. eCollection 2025 Apr. Hortic Res. 2024. PMID: 40070402 Free PMC article.
-
Recent Advances in Studying the Regulation of Fruit Ripening in Tomato Using Genetic Engineering Approaches.Int J Mol Sci. 2024 Jan 7;25(2):760. doi: 10.3390/ijms25020760. Int J Mol Sci. 2024. PMID: 38255834 Free PMC article. Review.
-
Expression of Vitis amurensis VaERF20 in Arabidopsis thaliana Improves Resistance to Botrytis cinerea and Pseudomonas syringae pv. Tomato DC3000.Int J Mol Sci. 2018 Mar 1;19(3):696. doi: 10.3390/ijms19030696. Int J Mol Sci. 2018. PMID: 29494485 Free PMC article.
-
The transcription factors VaERF16 and VaMYB306 interact to enhance resistance of grapevine to Botrytis cinerea infection.Mol Plant Pathol. 2022 Oct;23(10):1415-1432. doi: 10.1111/mpp.13223. Epub 2022 Jul 12. Mol Plant Pathol. 2022. PMID: 35822262 Free PMC article.
References
-
- Baxter, A. , Mittler, R. and Suzuki, N. (2014) ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. - PubMed
-
- Bombarely, A. , Rosli, H.G. , Vrebalov, J. , Moffett, P. , Mueller, L.A. and Martin, G.B. (2012) A draft genome sequence of Nicotiana benthamiana to enhance molecular plant–microbe biology research. Mol. Plant–Microbe Interact. 25, 1523–1530. - PubMed
-
- Brommonschenkel, S.H. , Frary, A. , Frary, A. and Tanksley, S.D. (2000) The broad‐spectrum tospovirus resistance gene Sw‐5 of tomato is a homolog of the root‐knot nematode resistance gene Mi . Mol. Plant–Microbe Interact. 13, 1130–1138. - PubMed
-
- Chen, Y.Y. , Lin, Y.M. , Chao, T.C. , Wang, J.F. , Liu, A.C. , Ho, F.I. and Cheng, C.P. (2009) Virus‐induced gene silencing reveals the involvement of ethylene‐, salicylic acid‐ and mitogen‐activated protein kinase‐related defense pathways in the resistance of tomato to bacterial wilt. Physiol. Plant. 136, 324–335. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

