Eukaryotic replication origins: Strength in flexibility
- PMID: 27416360
- PMCID: PMC4991242
- DOI: 10.1080/19491034.2016.1187353
Eukaryotic replication origins: Strength in flexibility
Abstract
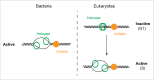
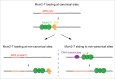
The eukaryotic replicative DNA helicase, Mcm2-7, is loaded in inactive form as a double hexameric complex around double-stranded DNA. To ensure that replication origins fire no more than once per S phase, activation of the Mcm2-7 helicase is temporally separated from Mcm2-7 loading in the cell cycle. This 2-step mechanism requires that inactive Mcm2-7 complexes be maintained for variable periods of time in a topologically bound state on chromatin, which may create a steric obstacle to other DNA transactions. We have recently found in the budding yeast, Saccharomyces cerevisiae, that Mcm2-7 double hexamers can respond to collisions with transcription complexes by sliding along the DNA template. Importantly, Mcm2-7 double hexamers remain functional after displacement along DNA and support replication initiation from sites distal to the origin. These results reveal a novel mechanism to specify eukaryotic replication origin sites and to maintain replication origin competence without the need for Mcm2-7 reloading.
Keywords: DNA replication; Mcm2-7; ORC; chromosomes; replication origin.
Figures
Comment on
- doi: 10.1016/j.molcel.2015.10.022
Similar articles
-
Post-licensing Specification of Eukaryotic Replication Origins by Facilitated Mcm2-7 Sliding along DNA.Mol Cell. 2015 Dec 3;60(5):797-807. doi: 10.1016/j.molcel.2015.10.022. Epub 2015 Nov 19. Mol Cell. 2015. PMID: 26656162 Free PMC article.
-
Replication origin-flanking roadblocks reveal origin-licensing dynamics and altered sequence dependence.J Biol Chem. 2017 Dec 29;292(52):21417-21430. doi: 10.1074/jbc.M117.815639. Epub 2017 Oct 26. J Biol Chem. 2017. PMID: 29074622 Free PMC article.
-
Regulation of replication origin licensing by ORC phosphorylation reveals a two-step mechanism for Mcm2-7 ring closing.Proc Natl Acad Sci U S A. 2023 Jul 18;120(29):e2221484120. doi: 10.1073/pnas.2221484120. Epub 2023 Jul 10. Proc Natl Acad Sci U S A. 2023. PMID: 37428921 Free PMC article.
-
The eukaryotic Mcm2-7 replicative helicase.Subcell Biochem. 2012;62:113-34. doi: 10.1007/978-94-007-4572-8_7. Subcell Biochem. 2012. PMID: 22918583 Review.
-
Switch on the engine: how the eukaryotic replicative helicase MCM2-7 becomes activated.Chromosoma. 2015 Mar;124(1):13-26. doi: 10.1007/s00412-014-0489-2. Epub 2014 Oct 12. Chromosoma. 2015. PMID: 25308420 Review.
Cited by
-
From structure to mechanism-understanding initiation of DNA replication.Genes Dev. 2017 Jun 1;31(11):1073-1088. doi: 10.1101/gad.298232.117. Genes Dev. 2017. PMID: 28717046 Free PMC article. Review.
-
Mistimed origin licensing and activation stabilize common fragile sites under tight DNA-replication checkpoint activation.Nat Struct Mol Biol. 2023 Apr;30(4):539-550. doi: 10.1038/s41594-023-00949-1. Epub 2023 Apr 6. Nat Struct Mol Biol. 2023. PMID: 37024657
-
Episomal and chromosomal DNA replication and recombination in Entamoeba histolytica.Front Mol Biosci. 2023 Jun 8;10:1212082. doi: 10.3389/fmolb.2023.1212082. eCollection 2023. Front Mol Biosci. 2023. PMID: 37363402 Free PMC article. Review.
-
Disruption of Toxoplasma gondii-Induced Host Cell DNA Replication Is Dependent on Contact Inhibition and Host Cell Type.mSphere. 2022 Jun 29;7(3):e0016022. doi: 10.1128/msphere.00160-22. Epub 2022 May 19. mSphere. 2022. PMID: 35587658 Free PMC article.
-
Regulation of DNA Replication Licensing and Re-Replication by Cdt1.Int J Mol Sci. 2021 May 14;22(10):5195. doi: 10.3390/ijms22105195. Int J Mol Sci. 2021. PMID: 34068957 Free PMC article. Review.
References
-
- Gaillard H, Aguilera A. Transcription as a Threat to Genome Integrity. Ann Rev Biochem 2016; PMID:27023844 - PubMed
-
- Siddiqui K, On KF, Diffley JF. Regulating DNA replication in eukarya. Cold Spring Harb Perspect Biol 2013; 5; PMID:23838438; http://dx.doi.org/10.1101/cshperspect.a012930 - DOI - PMC - PubMed
-
- Remus D, Diffley JF. Eukaryotic DNA replication control: lock and load, then fire. Curr Opin Cell Biol 2009; 21:771-7; PMID:19767190; http://dx.doi.org/10.1016/j.ceb.2009.08.002 - DOI - PubMed
-
- Rhind N, Gilbert DM. DNA replication timing. Cold Spring Harb Perspect Biol 2013; 5:a010132; PMID:23838440; http://dx.doi.org/10.1101/cshperspect.a010132 - DOI - PMC - PubMed
-
- Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci U S A 1997; 94:5611-6; PMID:9159120; http://dx.doi.org/10.1073/pnas.94.11.5611 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
