Assembling of the Mycobacterium tuberculosis Cell Wall Core
- PMID: 27417139
- PMCID: PMC5009262
- DOI: 10.1074/jbc.M116.739227
Assembling of the Mycobacterium tuberculosis Cell Wall Core
Abstract
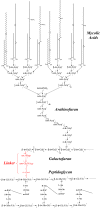
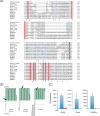
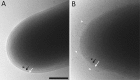
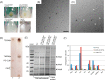
The unique cell wall of mycobacteria is essential to their viability and the target of many clinically used anti-tuberculosis drugs and inhibitors under development. Despite intensive efforts to identify the ligase(s) responsible for the covalent attachment of the two major heteropolysaccharides of the mycobacterial cell wall, arabinogalactan (AG) and peptidoglycan (PG), the enzyme or enzymes responsible have remained elusive. We here report on the identification of the two enzymes of Mycobacterium tuberculosis, CpsA1 (Rv3267) and CpsA2 (Rv3484), responsible for this function. CpsA1 and CpsA2 belong to the widespread LytR-Cps2A-Psr (LCP) family of enzymes that has been shown to catalyze a variety of glycopolymer transfer reactions in Gram-positive bacteria, including the attachment of wall teichoic acids to PG. Although individual cpsA1 and cpsA2 knock-outs of M. tuberculosis were readily obtained, the combined inactivation of both genes appears to be lethal. In the closely related microorganism Corynebacterium glutamicum, the ortholog of cpsA1 is the only gene involved in this function, and its conditional knockdown leads to dramatic changes in the cell wall composition and morphology of the bacteria due to extensive shedding of cell wall material in the culture medium as a result of defective attachment of AG to PG. This work marks an important step in our understanding of the biogenesis of the unique cell envelope of mycobacteria and opens new opportunities for drug development.
Keywords: Mycobacterium tuberculosis; arabinogalactan; cell wall; cryo-electron microscopy; ligase; outer membrane; peptidoglycan; polysaccharide.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Daffé M., and Draper P. (1998) The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39, 131–203 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

