Septin 9 induces lipid droplets growth by a phosphatidylinositol-5-phosphate and microtubule-dependent mechanism hijacked by HCV
- PMID: 27417143
- PMCID: PMC4947189
- DOI: 10.1038/ncomms12203
Septin 9 induces lipid droplets growth by a phosphatidylinositol-5-phosphate and microtubule-dependent mechanism hijacked by HCV
Abstract
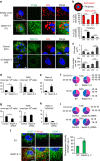
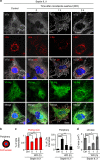
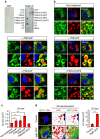
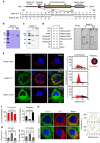
The accumulation of lipid droplets (LD) is frequently observed in hepatitis C virus (HCV) infection and represents an important risk factor for the development of liver steatosis and cirrhosis. The mechanisms of LD biogenesis and growth remain open questions. Here, transcriptome analysis reveals a significant upregulation of septin 9 in HCV-induced cirrhosis compared with the normal liver. HCV infection increases septin 9 expression and induces its assembly into filaments. Septin 9 regulates LD growth and perinuclear accumulation in a manner dependent on dynamic microtubules. The effects of septin 9 on LDs are also dependent on binding to PtdIns5P, which, in turn, controls the formation of septin 9 filaments and its interaction with microtubules. This previously undescribed cooperation between PtdIns5P and septin 9 regulates oleate-induced accumulation of LDs. Overall, our data offer a novel route for LD growth through the involvement of a septin 9/PtdIns5P signalling pathway.
Figures





References
-
- Ohsaki Y., Suzuki M. & Fujimoto T. Open questions in lipid droplet biology. Chem. Biol. 21, 86–96 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

