Challenges in identifying cancer genes by analysis of exome sequencing data
- PMID: 27417679
- PMCID: PMC4947162
- DOI: 10.1038/ncomms12096
Challenges in identifying cancer genes by analysis of exome sequencing data
Abstract
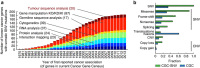
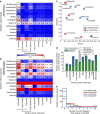
Massively parallel sequencing has permitted an unprecedented examination of the cancer exome, leading to predictions that all genes important to cancer will soon be identified by genetic analysis of tumours. To examine this potential, here we evaluate the ability of state-of-the-art sequence analysis methods to specifically recover known cancer genes. While some cancer genes are identified by analysis of recurrence, spatial clustering or predicted impact of somatic mutations, many remain undetected due to lack of power to discriminate driver mutations from the background mutational load (13-60% recall of cancer genes impacted by somatic single-nucleotide variants, depending on the method). Cancer genes not detected by mutation recurrence also tend to be missed by all types of exome analysis. Nonetheless, these genes are implicated by other experiments such as functional genetic screens and expression profiling. These challenges are only partially addressed by increasing sample size and will likely hold even as greater numbers of tumours are analysed.
Figures