Right on Q: genetics begin to unravel Coxiella burnetii host cell interactions
- PMID: 27418426
- PMCID: PMC5619019
- DOI: 10.2217/fmb-2016-0044
Right on Q: genetics begin to unravel Coxiella burnetii host cell interactions
Abstract
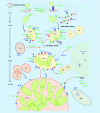
Invasion of macrophages and replication within an acidic and degradative phagolysosome-like vacuole are essential for disease pathogenesis by Coxiella burnetii, the bacterial agent of human Q fever. Previous experimental constraints imposed by the obligate intracellular nature of Coxiella limited knowledge of pathogen strategies that promote infection. Fortunately, new genetic tools facilitated by axenic culture now allow allelic exchange and transposon mutagenesis approaches for virulence gene discovery. Phenotypic screens have illuminated the critical importance of Coxiella's type 4B secretion system in host cell subversion and discovered genes encoding translocated effector proteins that manipulate critical infection events. Here, we highlight the cellular microbiology and genetics of Coxiella and how recent technical advances now make Coxiella a model organism to study macrophage parasitism.
Keywords: Coxiella burnetii; apoptosis; autophagy; effector protein; host cell invasion; macrophages; mutagenesis; phagolysosome; type 4B secretion; vacuole remodeling.
Conflict of interest statement
Figures
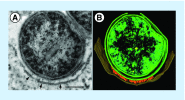
References
-
- Raoult D, Marrie T, Mege J. Natural history and pathophysiology of Q fever. Lancet Infect. Dis. 2005;5:219–226. - PubMed
-
- Babudieri C. Q fever: a zoonosis. Adv. Vet. Sci. 1959;5:81–84.
-
- Angelakis E, Raoult D. Q fever. Vet. Microbiol. 2010;140:297–309. - PubMed
-
- Hackert VH, Van Der Hoek W, Dukers-Muijrers N, et al. Q fever: single-point source outbreak with high attack rates and massive numbers of undetected infections across an entire region. Clin. Infect. Dis. 2012;55:1591–1599. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
