Utilization of Whole Exome Sequencing to Identify Causative Mutations in Familial Congenital Heart Disease
- PMID: 27418595
- PMCID: PMC5412122
- DOI: 10.1161/CIRCGENETICS.115.001324
Utilization of Whole Exome Sequencing to Identify Causative Mutations in Familial Congenital Heart Disease
Abstract
Background: Congenital heart disease (CHD) is the most common type of birth defect with family- and population-based studies supporting a strong genetic cause for CHD. The goal of this study was to determine whether a whole exome sequencing (WES) approach could identify pathogenic-segregating variants in multiplex CHD families.
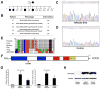
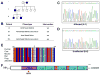
Methods and results: WES was performed on 9 kindreds with familial CHD, 4 with atrial septal defects, 2 with patent ductus arteriosus, 2 with tetralogy of Fallot, and 1 with pulmonary valve dysplasia. Rare variants (<1% minor allele frequency) that segregated with disease were identified by WES, and variants in 69 CHD candidate genes were further analyzed. These selected variants were subjected to in silico analysis to predict pathogenicity and resulted in the discovery of likely pathogenic mutations in 3 of 9 (33%) families. A GATA4 mutation in the transactivation domain, p.G115W, was identified in familial atrial septal defects and demonstrated decreased transactivation ability in vitro. A p.I263V mutation in TLL1 was identified in an atrial septal defects kindred and is predicted to affect the enzymatic functionality of TLL1. A disease-segregating splice donor site mutation in MYH11 (c.4599+1delG) was identified in familial patent ductus arteriosus and found to disrupt normal splicing of MYH11 mRNA in the affected individual.
Conclusions: Our findings demonstrate the clinical utility of WES to identify causative mutations in familial CHD and demonstrate the successful use of a CHD candidate gene list to allow for a more streamlined approach enabling rapid prioritization and identification of likely pathogenic variants from large WES data sets.
Clinical trial registration: URL: https://clinicaltrials.gov; Unique Identifier: NCT0112048.
Keywords: ductus arteriosus, patent; exome; genetic testing; heart septal defects, atrial; tetralogy of Fallot.
© 2016 American Heart Association, Inc.
Figures

Comment in
-
The Heritable Basis of Congenital Heart Disease: Past, Present, and Future.Circ Cardiovasc Genet. 2016 Aug;9(4):315-7. doi: 10.1161/CIRCGENETICS.116.001559. Circ Cardiovasc Genet. 2016. PMID: 27531915 Free PMC article. No abstract available.
Similar articles
-
The M310T mutation in the GATA4 gene is a novel pathogenic target of the familial atrial septal defect.BMC Cardiovasc Disord. 2021 Jan 6;21(1):12. doi: 10.1186/s12872-020-01822-5. BMC Cardiovasc Disord. 2021. PMID: 33413087 Free PMC article.
-
TBX20 loss-of-function mutation responsible for familial tetralogy of Fallot or sporadic persistent truncus arteriosus.Int J Med Sci. 2017 Mar 11;14(4):323-332. doi: 10.7150/ijms.17834. eCollection 2017. Int J Med Sci. 2017. PMID: 28553164 Free PMC article.
-
Whole-exome sequencing identifies R1279X of MYH6 gene to be associated with congenital heart disease.BMC Cardiovasc Disord. 2018 Jul 3;18(1):137. doi: 10.1186/s12872-018-0867-4. BMC Cardiovasc Disord. 2018. PMID: 29969989 Free PMC article.
-
Congenital heart diseases and their association with the variant distribution features on susceptibility genes.Clin Genet. 2017 Mar;91(3):349-354. doi: 10.1111/cge.12835. Epub 2016 Sep 5. Clin Genet. 2017. PMID: 27426723 Review.
-
Molecular determinants of atrial and ventricular septal defects and patent ductus arteriosus.Am J Med Genet. 2000 Winter;97(4):304-9. doi: 10.1002/1096-8628(200024)97:4<304::aid-ajmg1281>3.0.co;2-#. Am J Med Genet. 2000. PMID: 11376442 Review.
Cited by
-
Genomic Risk Factors Driving Immune-Mediated Delayed Drug Hypersensitivity Reactions.Front Genet. 2021 Apr 16;12:641905. doi: 10.3389/fgene.2021.641905. eCollection 2021. Front Genet. 2021. PMID: 33936169 Free PMC article. Review.
-
Genetic Basis for Congenital Heart Disease: Revisited: A Scientific Statement From the American Heart Association.Circulation. 2018 Nov 20;138(21):e653-e711. doi: 10.1161/CIR.0000000000000606. Circulation. 2018. PMID: 30571578 Free PMC article. Review.
-
Role of Segregation for Variant Discovery in Multiplex Families Ascertained by Probands With Left Sided Cardiovascular Malformations.Front Genet. 2019 Jan 11;9:729. doi: 10.3389/fgene.2018.00729. eCollection 2018. Front Genet. 2019. PMID: 30687393 Free PMC article.
-
Genetics and Genomics of Congenital Heart Disease.Circ Res. 2017 Mar 17;120(6):923-940. doi: 10.1161/CIRCRESAHA.116.309140. Circ Res. 2017. PMID: 28302740 Free PMC article. Review.
-
Single-cell reconstruction and mutation enrichment analysis identifies dysregulated cardiomyocyte and endothelial cells in congenital heart disease.Physiol Genomics. 2023 Dec 1;55(12):634-646. doi: 10.1152/physiolgenomics.00070.2023. Epub 2023 Oct 9. Physiol Genomics. 2023. PMID: 37811720 Free PMC article.
References
-
- Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–1900. - PubMed
-
- Marelli AJ, Mackie AS, Ionescu-Ittu R, Rahme E, Pilote L. Congenital heart disease in the general population: changing prevalence and age distribution. Circulation. 2007;115:163–172. - PubMed
-
- Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. - PubMed
-
- Miller A, Riehle-Colarusso T, Alverson CJ, Frias JL, Correa A. Congenital heart defects and major structural noncardiac anomalies, Atlanta, Georgia, 1968 to 2005. J Pediatr. 2011;159:70–78. e2. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

