On-Chip Clonal Analysis of Glioma-Stem-Cell Motility and Therapy Resistance
- PMID: 27420544
- PMCID: PMC5040341
- DOI: 10.1021/acs.nanolett.6b00902
On-Chip Clonal Analysis of Glioma-Stem-Cell Motility and Therapy Resistance
Abstract
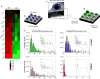
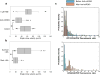
Enhanced glioma-stem-cell (GSC) motility and therapy resistance are considered to play key roles in tumor cell dissemination and recurrence. As such, a better understanding of the mechanisms by which these cells disseminate and withstand therapy could lead to more efficacious treatments. Here, we introduce a novel micro-/nanotechnology-enabled chip platform for performing live-cell interrogation of patient-derived GSCs with single-clone resolution. On-chip analysis revealed marked intertumoral differences (>10-fold) in single-clone motility profiles between two populations of GSCs, which correlated well with results from tumor-xenograft experiments and gene-expression analyses. Further chip-based examination of the more-aggressive GSC population revealed pronounced interclonal variations in motility capabilities (up to ∼4-fold) as well as gene-expression profiles at the single-cell level. Chip-supported therapy resistance studies with a chemotherapeutic agent (i.e., temozolomide) and an oligo RNA (anti-miR363) revealed a subpopulation of CD44-high GSCs with strong antiapoptotic behavior as well as enhanced motility capabilities. The living-cell-interrogation chip platform described herein enables thorough and large-scale live monitoring of heterogeneous cancer-cell populations with single-cell resolution, which is not achievable by any other existing technology and thus has the potential to provide new insights into the cellular and molecular mechanisms modulating glioma-stem-cell dissemination and therapy resistance.
Keywords: Living single-cell analysis; anti-microRNA; cancer stem cell; cell motility; glioblastoma; nanochannel electroporation.
Conflict of interest statement
Notes The authors declare no competing financial interest.
Figures


References
-
- Bellail AC, Hunter SB, Brat DJ, Tan C, Van Meir EG. Int J Biochem Cell Biol. 2004;36(6):1046–69. - PubMed
-
- Gallego-Perez D, Higuita-Castro N, Denning L, DeJesus J, Dahl K, Sarkar A, Hansford DJ. Lab Chip. 2012;12(21):4424–32. - PubMed
-
- Giese A, Bjerkvig R, Berens ME, Westphal M. J Clin Oncol. 2003;21(8):1624–36. - PubMed
-
- Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, Frank A, Bayazitov IT, Zakharenko SS, Gajjar A, Davidoff A, Gilbertson RJ. Cancer Cell. 2007;11(1):69–82. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

