Characterization of a potential ABC-type bacteriocin exporter protein from Treponema denticola
- PMID: 27422166
- PMCID: PMC4947327
- DOI: 10.1186/s12903-016-0243-7
Characterization of a potential ABC-type bacteriocin exporter protein from Treponema denticola
Abstract
Background: Treponema denticola is strongly associated with the development of periodontal disease. Both synergistic and antagonistic effects are observed among bacterial species in the process of biofilm formation. Bacteriocin-related genes have not yet been fully characterized in periodontopathic bacteria. The aim of this study was to detect and characterize bacteriocin-associated proteins in T. denticola.
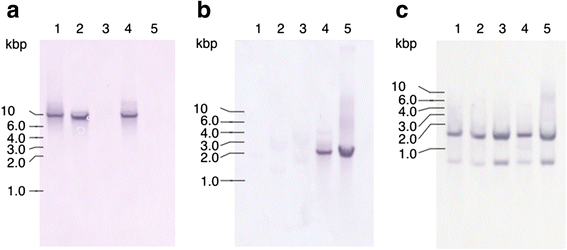
Methods: The whole genome sequence of T. denticola ATCC 35405 was screened with a Streptococcus mutans bacteriocin immunity protein (ImmA/Bip) sequence. The prevalence of homologous genes in T. denticola strains was then investigated by Southern blotting. Expression of the genes was evaluated by qRT-PCR.
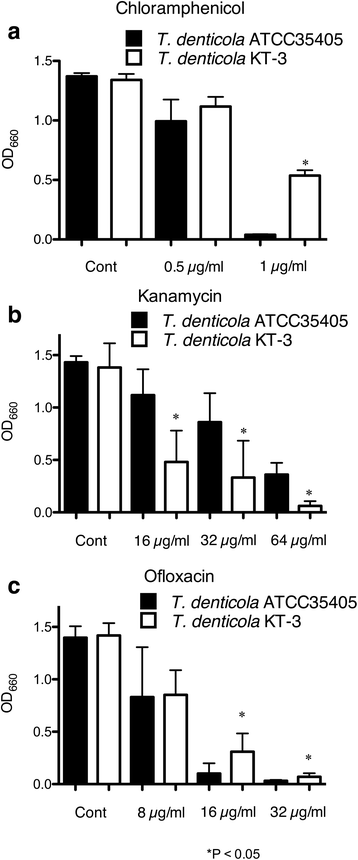
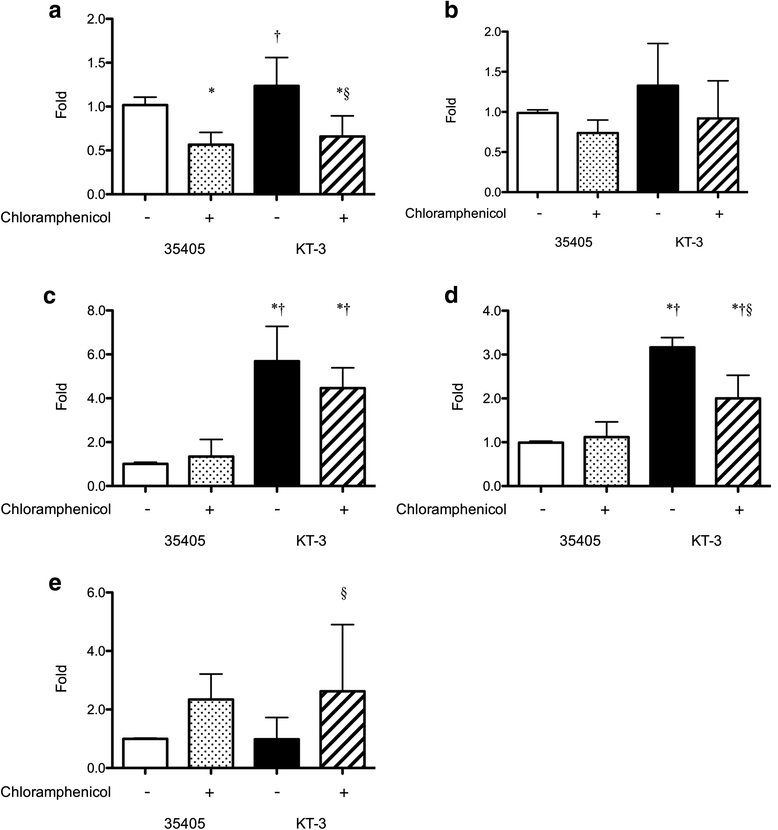
Results: In the genome sequence of T. denticola, an amino acid sequence coded by the open reading frame TDE_0719 showed 26 % identity with the S. mutans ImmA. Furthermore, two protein sequences encoded by TDE_0425 and TDE_2431 in T. denticola ATCC 35405 showed ~40 % identity with that coded by TDE_0719. Therefore, TDE_0425, TDE_0719, and TDE_2431 were designated as tepA1, A2, and A3, respectively. Open reading frames showing similarity to the HlyD family of secretion proteins were detected downstream of tepA1, A2, and A3. They were designated as tepB1, B2, and B3, respectively. A gene harboring a bacteriocin-like signal sequence was detected upstream of tepA1. The prevalence of tepA1 and A2 differed among Treponema species. Susceptibility to chloramphenicol and ofloxacin was slightly decreased in a tepA2 mutant while that to kanamycin was increased. Expression of tepA3-B3 was increased in the tepA2 mutant.
Conclusion: These results indicate that T. denticola ATCC 35405 has three potential bacteriocin export proteins and that the presence of these genes differs among the Treponema strains. TepA3-B3 of the corresponding proteins may be involved in resistance to chloramphenicol.
Keywords: ABC transporter; Antimicrobial agent susceptibility; Bacteriocin; Treponema denticola.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

