Enrichment of Root Endophytic Bacteria from Populus deltoides and Single-Cell-Genomics Analysis
- PMID: 27422831
- PMCID: PMC5007785
- DOI: 10.1128/AEM.01285-16
Enrichment of Root Endophytic Bacteria from Populus deltoides and Single-Cell-Genomics Analysis
Abstract
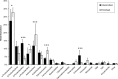
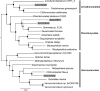
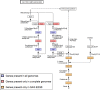
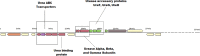
Bacterial endophytes that colonize Populus trees contribute to nutrient acquisition, prime immunity responses, and directly or indirectly increase both above- and below-ground biomasses. Endophytes are embedded within plant material, so physical separation and isolation are difficult tasks. Application of culture-independent methods, such as metagenome or bacterial transcriptome sequencing, has been limited due to the predominance of DNA from the plant biomass. Here, we describe a modified differential and density gradient centrifugation-based protocol for the separation of endophytic bacteria from Populus roots. This protocol achieved substantial reduction in contaminating plant DNA, allowed enrichment of endophytic bacteria away from the plant material, and enabled single-cell genomics analysis. Four single-cell genomes were selected for whole-genome amplification based on their rarity in the microbiome (potentially uncultured taxa) as well as their inferred abilities to form associations with plants. Bioinformatics analyses, including assembly, contamination removal, and completeness estimation, were performed to obtain single-amplified genomes (SAGs) of organisms from the phyla Armatimonadetes, Verrucomicrobia, and Planctomycetes, which were unrepresented in our previous cultivation efforts. Comparative genomic analysis revealed unique characteristics of each SAG that could facilitate future cultivation efforts for these bacteria.
Importance: Plant roots harbor a diverse collection of microbes that live within host tissues. To gain a comprehensive understanding of microbial adaptations to this endophytic lifestyle from strains that cannot be cultivated, it is necessary to separate bacterial cells from the predominance of plant tissue. This study provides a valuable approach for the separation and isolation of endophytic bacteria from plant root tissue. Isolated live bacteria provide material for microbiome sequencing, single-cell genomics, and analyses of genomes of uncultured bacteria to provide genomics information that will facilitate future cultivation attempts.
Copyright © 2016 Utturkar et al.
Figures
References
-
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi:10.1038/nature12352. - DOI - PubMed
-
- Seshadri R, Paulsen IT, Eisen JA, Read TD, Nelson KE, Nelson WC, Ward NL, Tettelin H, Davidsen TM, Beanan MJ, Deboy RT, Daugherty SC, Brinkac LM, Madupu R, Dodson RJ, Khouri HM, Lee KH, Carty HA, Scanlan D, Heinzen RA, Thompson HA, Samuel JE, Fraser CM, Heidelberg JF. 2003. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc Natl Acad Sci U S A 100:5455–5460. doi:10.1073/pnas.0931379100. - DOI - PMC - PubMed
-
- Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schaberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K. 2015. A new antibiotic kills pathogens without detectable resistance. Nature 517:455–459. doi:10.1038/nature14098. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

