Lactobacillus rhamnosus GG Outcompetes Enterococcus faecium via Mucus-Binding Pili: Evidence for a Novel and Heterospecific Probiotic Mechanism
- PMID: 27422834
- PMCID: PMC5038030
- DOI: 10.1128/AEM.01243-16
Lactobacillus rhamnosus GG Outcompetes Enterococcus faecium via Mucus-Binding Pili: Evidence for a Novel and Heterospecific Probiotic Mechanism
Abstract
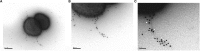
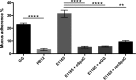
Vancomycin-resistant enterococci (VRE) have become a major nosocomial threat. Enterococcus faecium is of special concern, as it can easily acquire new antibiotic resistances and is an excellent colonizer of the human intestinal tract. Several clinical studies have explored the potential use of beneficial bacteria to weed out opportunistic pathogens. Specifically, the widely studied Lactobacillus rhamnosus strain GG has been applied successfully in the context of VRE infections. Here, we provide new insight into the molecular mechanism underlying the effects of this model probiotic on VRE decolonization. Both clinical VRE isolates and L. rhamnosus GG express pili on their cell walls, which are the key modulators of their highly efficient colonization of the intestinal mucosa. We found that one of the VRE pilus clusters shares considerable sequence similarity with the SpaCBA-SrtC1 pilus cluster of L. rhamnosus GG. Remarkable immunological and functional similarities were discovered between the mucus-binding pili of L. rhamnosus GG and those of the clinical E. faecium strain E1165, which was characterized at the genome level. Moreover, E. faecium strain E1165 bound efficiently to mucus, which may be prevented by the presence of the mucus-binding SpaC protein or antibodies against L. rhamnosus GG or SpaC. These results present experimental support for a novel probiotic mechanism, in which the mucus-binding pili of L. rhamnosus GG prevent the binding of a potential pathogen to the host. Hence, we provide a molecular basis for the further exploitation of L. rhamnosus GG and its pilins for prophylaxis and treatment of VRE infections.
Importance: Concern about vancomycin-resistant Enterococcus faecium causing nosocomial infections is rising globally. The arsenal of antibiotic strategies to treat these infections is nearly exhausted, and hence, new treatment strategies are urgently needed. Here, we provide molecular evidence to underpin reports of the successful clinical application of Lactobacillus rhamnosus GG in VRE decolonization strategies. Our results provide support for a new molecular mechanism, in which probiotics can perform competitive exclusion and possibly immune interaction. Moreover, we spur further exploration of the potential of intact L. rhamnosus GG and purified SpaC pilin as prophylactic and curative agents of the VRE carrier state.
Copyright © 2016 Tytgat et al.
Figures
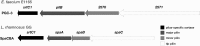


Similar articles
-
Characterization of the SpaCBA pilus fibers in the probiotic Lactobacillus rhamnosus GG.Appl Environ Microbiol. 2012 Apr;78(7):2337-44. doi: 10.1128/AEM.07047-11. Epub 2012 Jan 13. Appl Environ Microbiol. 2012. PMID: 22247175 Free PMC article.
-
Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein.Proc Natl Acad Sci U S A. 2009 Oct 6;106(40):17193-8. doi: 10.1073/pnas.0908876106. Epub 2009 Sep 17. Proc Natl Acad Sci U S A. 2009. PMID: 19805152 Free PMC article.
-
Selection and characterization of a SpaCBA pilus-secreting food-grade derivative of Lacticaseibacillus rhamnosus GG.Appl Microbiol Biotechnol. 2021 Feb;105(3):1123-1131. doi: 10.1007/s00253-020-11051-7. Epub 2021 Jan 8. Appl Microbiol Biotechnol. 2021. PMID: 33417041 Free PMC article.
-
Decolonization of gastrointestinal carriage of vancomycin-resistant Enterococcus faecium: case series and review of literature.BMC Infect Dis. 2014 Sep 23;14:514. doi: 10.1186/1471-2334-14-514. BMC Infect Dis. 2014. PMID: 25248287 Free PMC article. Review.
-
Lactobacillus rhamnosus sepsis associated with probiotic therapy in an extremely preterm infant: Pathogenesis and a review for clinicians.J Microbiol Immunol Infect. 2021 Aug;54(4):575-580. doi: 10.1016/j.jmii.2020.03.029. Epub 2020 Apr 3. J Microbiol Immunol Infect. 2021. PMID: 32307246 Review.
Cited by
-
Enlisting commensal microbes to resist antibiotic-resistant pathogens.J Exp Med. 2019 Jan 7;216(1):10-19. doi: 10.1084/jem.20180399. Epub 2018 Oct 11. J Exp Med. 2019. PMID: 30309968 Free PMC article. Review.
-
Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health.Appl Microbiol Biotechnol. 2019 Aug;103(16):6463-6472. doi: 10.1007/s00253-019-09978-7. Epub 2019 Jul 2. Appl Microbiol Biotechnol. 2019. PMID: 31267231 Free PMC article. Review.
-
Anti-Infective Effect of Adhesive Probiotic Lactobacillus in Fish is Correlated With Their Spatial Distribution in the Intestinal Tissue.Sci Rep. 2017 Oct 16;7(1):13195. doi: 10.1038/s41598-017-13466-1. Sci Rep. 2017. PMID: 29038557 Free PMC article.
-
Colorectal Cancer Mitigation Through Probiotics: Current Evidence and Future Directions.Curr Microbiol. 2025 Jun 18;82(8):339. doi: 10.1007/s00284-025-04297-9. Curr Microbiol. 2025. PMID: 40533631 Review.
-
No Effect of Lactobacillus rhamnosus GG on Eradication of Colonization by Vancomycin-Resistant Enterococcus faecium or Microbiome Diversity in Hospitalized Adult Patients.Microbiol Spectr. 2022 Jun 29;10(3):e0234821. doi: 10.1128/spectrum.02348-21. Epub 2022 Apr 27. Microbiol Spectr. 2022. PMID: 35475684 Free PMC article. Clinical Trial.
References
-
- Uttley AH, Collins CH, Naidoo J, George RC. 1988. Vancomycin-resistant enterococci. Lancet i:57–58. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

