The Presence of Two Receptor-Binding Proteins Contributes to the Wide Host Range of Staphylococcal Twort-Like Phages
- PMID: 27422842
- PMCID: PMC5038044
- DOI: 10.1128/AEM.01385-16
The Presence of Two Receptor-Binding Proteins Contributes to the Wide Host Range of Staphylococcal Twort-Like Phages
Abstract
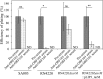
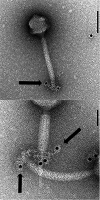
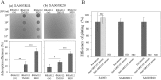
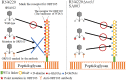
Thanks to their wide host range and virulence, staphylococcal bacteriophages (phages) belonging to the genus Twortlikevirus (staphylococcal Twort-like phages) are regarded as ideal candidates for clinical application for Staphylococcus aureus infections due to the emergence of antibiotic-resistant bacteria of this species. To increase the usability of these phages, it is necessary to understand the mechanism underlying host recognition, especially the receptor-binding proteins (RBPs) that determine host range. In this study, we found that the staphylococcal Twort-like phage ΦSA012 possesses at least two RBPs. Genomic analysis of five mutant phages of ΦSA012 revealed point mutations in orf103, in a region unique to staphylococcal Twort-like phages. Phages harboring mutated ORF103 could not infect S. aureus strains in which wall teichoic acids (WTAs) are glycosylated with α-N-acetylglucosamine (α-GlcNAc). A polyclonal antibody against ORF103 also inhibited infection by ΦSA012 in the presence of α-GlcNAc, suggesting that ORF103 binds to α-GlcNAc. In contrast, a polyclonal antibody against ORF105, a short tail fiber component previously shown to be an RBP, inhibited phage infection irrespective of the presence of α-GlcNAc. Immunoelectron microscopy indicated that ORF103 is a tail fiber component localized at the bottom of the baseplate. From these results, we conclude that ORF103 binds α-GlcNAc in WTAs, whereas ORF105, the primary RBP, is likely to bind the WTA backbone. These findings provide insight into the infection mechanism of staphylococcal Twort-like phages.
Importance: Staphylococcus phages belonging to the genus Twortlikevirus (called staphylococcal Twort-like phages) are considered promising agents for control of Staphylococcus aureus due to their wide host range and highly lytic capabilities. Although staphylococcal Twort-like phages have been studied widely for therapeutic purposes, the host recognition process of staphylococcal Twort-like phages remains unclear. This work provides new findings about the mechanisms of host recognition of the staphylococcal Twort-like phage ΦSA012. The details of the host recognition mechanism of ΦSA012 will allow us to analyze the mechanisms of infection and expand the utility of staphylococcal Twort-like phages for the control of S. aureus.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Analysis of phage resistance in Staphylococcus aureus SA003 reveals different binding mechanisms for the closely related Twort-like phages ɸSA012 and ɸSA039.Appl Microbiol Biotechnol. 2018 Oct;102(20):8963-8977. doi: 10.1007/s00253-018-9269-x. Epub 2018 Aug 4. Appl Microbiol Biotechnol. 2018. PMID: 30078137
-
Analysis host-recognition mechanism of staphylococcal kayvirus ɸSA039 reveals a novel strategy that protects Staphylococcus aureus against infection by Staphylococcus pseudintermedius Siphoviridae phages.Appl Microbiol Biotechnol. 2019 Aug;103(16):6809-6823. doi: 10.1007/s00253-019-09940-7. Epub 2019 Jun 25. Appl Microbiol Biotechnol. 2019. PMID: 31236618
-
Silviavirus phage ɸMR003 displays a broad host range against methicillin-resistant Staphylococcus aureus of human origin.Appl Microbiol Biotechnol. 2019 Sep;103(18):7751-7765. doi: 10.1007/s00253-019-10039-2. Epub 2019 Aug 6. Appl Microbiol Biotechnol. 2019. PMID: 31388727
-
Genomics of staphylococcal Twort-like phages--potential therapeutics of the post-antibiotic era.Adv Virus Res. 2012;83:143-216. doi: 10.1016/B978-0-12-394438-2.00005-0. Adv Virus Res. 2012. PMID: 22748811 Review.
-
Peculiarities of Staphylococcus aureus phages and their possible application in phage therapy.Appl Microbiol Biotechnol. 2019 Jun;103(11):4279-4289. doi: 10.1007/s00253-019-09810-2. Epub 2019 Apr 17. Appl Microbiol Biotechnol. 2019. PMID: 30997551 Review.
Cited by
-
Interplays of mutations in waaA, cmk, and ail contribute to phage resistance in Yersinia pestis.Front Cell Infect Microbiol. 2023 May 26;13:1174510. doi: 10.3389/fcimb.2023.1174510. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37305418 Free PMC article.
-
Deploying Viruses against Phytobacteria: Potential Use of Phage Cocktails as a Multifaceted Approach to Combat Resistant Bacterial Plant Pathogens.Viruses. 2022 Jan 18;14(2):171. doi: 10.3390/v14020171. Viruses. 2022. PMID: 35215763 Free PMC article. Review.
-
Deciphering the adsorption machinery of Deep-Blue and Vp4, two myophages targeting members of the Bacillus cereus group.J Virol. 2024 Sep 17;98(9):e0074524. doi: 10.1128/jvi.00745-24. Epub 2024 Aug 23. J Virol. 2024. PMID: 39177355 Free PMC article.
-
Predicting phage-host interactions via feature augmentation and regional graph convolution.Brief Bioinform. 2024 Nov 22;26(1):bbae672. doi: 10.1093/bib/bbae672. Brief Bioinform. 2024. PMID: 39727002 Free PMC article.
-
Selection of mutant Listeria phages under food-relevant conditions can enhance application potential.Appl Environ Microbiol. 2023 Oct 31;89(10):e0100723. doi: 10.1128/aem.01007-23. Epub 2023 Oct 6. Appl Environ Microbiol. 2023. PMID: 37800961 Free PMC article.
References
-
- Matsuzaki S, Yasuda M, Nishikawa H, Kuroda M, Ujihara T, Shuin T, Shen Y, Jin Z, Fujimoto S, Nasimuzzaman MD, Wakiguchi H, Sugihara S, Sugiura T, Koda S, Muraoka A, Imai S. 2003. Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage phi MR11. J Infect Dis 187:613–624. doi:10.1086/374001. - DOI - PubMed
-
- Synnott AJ, Kuang Y, Kurimoto M, Yamamichi K, Iwano H, Tanji Y. 2009. Isolation from sewage influent and characterization of novel Staphylococcus aureus bacteriophages with wide host ranges and potent lytic capabilities. Appl Environ Microbiol 75:4483–4490. doi:10.1128/AEM.02641-08. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

