Chopping and Changing: the Evolution of the Flavin-dependent Monooxygenases
- PMID: 27423402
- PMCID: PMC4981433
- DOI: 10.1016/j.jmb.2016.07.003
Chopping and Changing: the Evolution of the Flavin-dependent Monooxygenases
Abstract
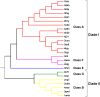
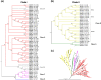
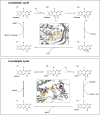
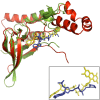
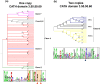
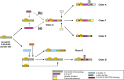
Flavin-dependent monooxygenases play a variety of key physiological roles and are also very powerful biotechnological tools. These enzymes have been classified into eight different classes (A-H) based on their sequences and biochemical features. By combining structural and sequence analysis, and phylogenetic inference, we have explored the evolutionary history of classes A, B, E, F, and G and demonstrate that their multidomain architectures reflect their phylogenetic relationships, suggesting that the main evolutionary steps in their divergence are likely to have arisen from the recruitment of different domains. Additionally, the functional divergence within in each class appears to have been the result of other mechanisms such as a complex set of single-point mutations. Our results reinforce the idea that a main constraint on the evolution of cofactor-dependent enzymes is the functional binding of the cofactor. Additionally, a remarkable feature of this family is that the sequence of the key flavin adenine dinucleotide-binding domain is split into at least two parts in all classes studied here. We propose a complex set of evolutionary events that gave rise to the origin of the different classes within this family.
Keywords: cofactor-binding enzymes; enzyme evolution; flavin-dependent monooxygenases; multidomain architecture.
Copyright © 2016 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures



References
-
- Macheroux P., Kappes B., Ealick S.E. Flavogenomics—a genomic and structural view of flavin-dependent proteins. FEBS J. 2011;278:2625–2634. - PubMed
-
- Mattevi A. To be or not to be an oxidase: challenging the oxygen reactivity of flavoenzymes. Trends Biochem. Sci. 2006;31:276–283. - PubMed
-
- Balke K., Kadow M., Mallin H., Sass S., Bornscheuer U.T. Discovery, application and protein engineering of Baeyer–Villiger monooxygenases for organic synthesis. Org. Biomol. Chem. 2012;10:6249–6265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

