Mutant IDH1 Downregulates ATM and Alters DNA Repair and Sensitivity to DNA Damage Independent of TET2
- PMID: 27424808
- PMCID: PMC5022794
- DOI: 10.1016/j.ccell.2016.05.018
Mutant IDH1 Downregulates ATM and Alters DNA Repair and Sensitivity to DNA Damage Independent of TET2
Abstract
Mutations in the isocitrate dehydrogenase-1 gene (IDH1) are common drivers of acute myeloid leukemia (AML) but their mechanism is not fully understood. It is thought that IDH1 mutants act by inhibiting TET2 to alter DNA methylation, but there are significant unexplained clinical differences between IDH1- and TET2-mutant diseases. We have discovered that mice expressing endogenous mutant IDH1 have reduced numbers of hematopoietic stem cells (HSCs), in contrast to Tet2 knockout (TET2-KO) mice. Mutant IDH1 downregulates the DNA damage (DD) sensor ATM by altering histone methylation, leading to impaired DNA repair, increased sensitivity to DD, and reduced HSC self-renewal, independent of TET2. ATM expression is also decreased in human IDH1-mutated AML. These findings may have implications for treatment of IDH-mutant leukemia.
Copyright © 2016 Elsevier Inc. All rights reserved.
Conflict of interest statement
Other authors declare that no conflicts of interest exist.
Figures


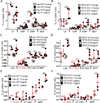


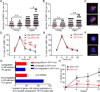


Comment in
-
IDH1, Histone Methylation, and So Forth.Cancer Cell. 2016 Aug 8;30(2):192-194. doi: 10.1016/j.ccell.2016.07.008. Cancer Cell. 2016. PMID: 27505668
References
-
- Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer discovery. 2013;3:730–741. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

