New insights into decapping enzymes and selective mRNA decay
- PMID: 27425147
- PMCID: PMC5179306
- DOI: 10.1002/wrna.1379
New insights into decapping enzymes and selective mRNA decay
Abstract
Removal of the 5' end cap is a critical determinant controlling mRNA stability and efficient gene expression. Removal of the cap is exquisitely controlled by multiple direct and indirect regulators that influence association with the cap and the catalytic step. A subset of these factors directly stimulate activity of the decapping enzyme, while others influence remodeling of factors bound to mRNA and indirectly stimulate decapping. Furthermore, the components of the general decapping machinery can also be recruited by mRNA-specific regulatory proteins to activate decapping. The Nudix hydrolase, Dcp2, identified as a first decapping enzyme, cleaves capped mRNA and initiates 5'-3' degradation. Extensive studies on Dcp2 led to broad understanding of its activity and the regulation of transcript specific decapping and decay. Interestingly, seven additional Nudix proteins possess intrinsic decapping activity in vitro and at least two, Nudt16 and Nudt3, are decapping enzymes that regulate mRNA stability in cells. Furthermore, a new class of decapping proteins within the DXO family preferentially function on incompletely capped mRNAs. Importantly, it is now evident that each of the characterized decapping enzymes predominantly modulates only a subset of mRNAs, suggesting the existence of multiple decapping enzymes functioning in distinct cellular pathways. WIREs RNA 2017, 8:e1379. doi: 10.1002/wrna.1379 For further resources related to this article, please visit the WIREs website.
© 2016 Wiley Periodicals, Inc.
Conflict of interest statement
statement. The authors have no conflict to declare.
Figures

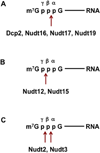
References
-
- Muthukrishnan S, Both GW, Furuichi Y, Shatkin AJ. 5'-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature. 1975;255:33–37. - PubMed
-
- Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA. 2011;2:277–298. - PubMed
-
- Konarska MM, Padgett RA, Sharp PA. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984;38:731–736. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

