Trichinella spiralis: Adaptation and parasitism
- PMID: 27425574
- PMCID: PMC5127757
- DOI: 10.1016/j.vetpar.2016.07.003
Trichinella spiralis: Adaptation and parasitism
Abstract
Publication of the genome from the clade I organism, Trichinella spiralis, has provided us an avenue to address more holistic problems in parasitology; namely the processes of adaptation and the evolution of parasitism. Parasitism among nematodes has evolved in multiple, independent events. Deciphering processes that drive species diversity and adaptation are keys to understanding parasitism and advancing control strategies. Studies have been put forth on morphological and physiological aspects of parasitism and adaptation in nematodes; however, data is now coming available to investigate adaptation, host switching and parasitism at the genomic level. Herein we compare proteomic data from the clade I parasite, Trichinella spiralis with data from Brugia malayi (clade III), Meloidogyne hapla and Meloidogyne incognita (clade IV), and free-living nematodes belonging to the genera Caenorhabditis and Pristionchus (clade V). We explore changes in protein family birth/death and expansion/reduction over the course of metazoan evolution using Homo sapiens, Drosophila melanogaster and Saccharomyces cerevisiae as outgroups for the phylum Nematoda. We further examine relationships between these changes and the ability and/or result of nematodes adapting to their environments. Data are consistent with gene loss occurring in conjunction with nematode specialization resulting from parasitic worms acclimating to well-defined, environmental niches. We observed evidence for independent, lateral gene transfer events involving conserved genes that may have played a role in the evolution of nematode parasitism. In general, parasitic nematodes gained proteins through duplication and lateral gene transfer, and lost proteins through random mutation and deletions. Data suggest independent acquisition rather than ancestral inheritance among the Nematoda followed by selective gene loss over evolutionary time. Data also show that parasitism and adaptation affected a broad range of proteins, especially those involved in sensory perception, metabolism, and transcription/translation. New protein gains with functions related to regulating transcription and translation, and protein family expansions with functions related to morphology and body development have occurred in association with parasitism. Further gains occurred as a result of lateral gene transfer and in particular, with the cyanase protein family In contrast, reductions and/or losses have occurred in protein families with functions related to metabolic process and signal transduction. Taking advantage of the independent occurrences of parasitism in nematodes, which enabled us to distinguish changes associated with parasitism from species specific niche adaptation, our study provides valuable insights into nematode parasitism at a proteome level using T. spiralis as a benchmark for early adaptation to or acquisition of parasitism.
Keywords: Adaptation; Brugia; Caenorhabditis; Genomics; Meloidogyne; Parasitism; Proteomics; Trichinella.
Published by Elsevier B.V.
Figures

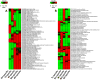
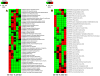




Similar articles
-
Comparative Genomic Analysis of Trichinella spiralis Reveals Potential Mechanisms of Adaptive Evolution.Biomed Res Int. 2019 May 21;2019:2948973. doi: 10.1155/2019/2948973. eCollection 2019. Biomed Res Int. 2019. PMID: 31240209 Free PMC article.
-
A tale of three kingdoms: members of the Phylum Nematoda independently acquired the detoxifying enzyme cyanase through horizontal gene transfer from plants and bacteria.Parasitology. 2019 Apr;146(4):445-452. doi: 10.1017/S0031182018001701. Epub 2018 Oct 10. Parasitology. 2019. PMID: 30301483 Free PMC article.
-
The draft genome of the parasitic nematode Trichinella spiralis.Nat Genet. 2011 Mar;43(3):228-35. doi: 10.1038/ng.769. Epub 2011 Feb 20. Nat Genet. 2011. PMID: 21336279 Free PMC article.
-
Advances in the sequencing of the genome of the adenophorean nematode Trichinella spiralis.Parasitology. 2008 Jul;135(8):869-80. doi: 10.1017/S0031182008004472. Parasitology. 2008. PMID: 18598573 Free PMC article. Review.
-
Trichinella spiralis: genomic application to control a zoonotic nematode.Infect Disord Drug Targets. 2010 Oct;10(5):376-84. doi: 10.2174/187152610793180830. Infect Disord Drug Targets. 2010. PMID: 20701572 Review.
Cited by
-
Comparative Genomic Analysis of Trichinella spiralis Reveals Potential Mechanisms of Adaptive Evolution.Biomed Res Int. 2019 May 21;2019:2948973. doi: 10.1155/2019/2948973. eCollection 2019. Biomed Res Int. 2019. PMID: 31240209 Free PMC article.
-
A tale of three kingdoms: members of the Phylum Nematoda independently acquired the detoxifying enzyme cyanase through horizontal gene transfer from plants and bacteria.Parasitology. 2019 Apr;146(4):445-452. doi: 10.1017/S0031182018001701. Epub 2018 Oct 10. Parasitology. 2019. PMID: 30301483 Free PMC article.
-
Trichinellosis-Induced Eosinophilic Myocarditis Mimicking Hypereosinophilic Syndrome.Cureus. 2024 Apr 24;16(4):e58946. doi: 10.7759/cureus.58946. eCollection 2024 Apr. Cureus. 2024. PMID: 38800259 Free PMC article.
-
Horizontal gene transfer provides insights into the deep evolutionary history and biology of Trichinella.Food Waterborne Parasitol. 2022 Apr 18;27:e00155. doi: 10.1016/j.fawpar.2022.e00155. eCollection 2022 Jun. Food Waterborne Parasitol. 2022. PMID: 35542181 Free PMC article.
-
Genotyping and Phylogenetic Position of Trichinella spiralis Isolates from Different Geographical Locations in China.Front Genet. 2019 Oct 31;10:1093. doi: 10.3389/fgene.2019.01093. eCollection 2019. Front Genet. 2019. PMID: 31737057 Free PMC article.
References
-
- Abad P, Gouzy J, Aury JM, Castagnone-Sereno P, Danchin EGJ, Deleury E, Perfus-Barbeoch L, Anthouard V, Artiguenave F, Blok VC, Caillaud MC, Coutinho PM, Dasilva C, De Luca F, Deau F, Esquibet M, Flutre T, Goldstone JV, Hamamouch N, Hewezi T, Jaillon O, Jubin C, Leonetti P, Magliano M, Maier TR, Markov GV, McVeigh P, Pesole G, Poulain J, Robinson-Rechavi M, Sallet E, Segurens B, Steinbach D, Tytgat T, Ugarte E, van Ghelder C, Veronico P, Baum TJ, Blaxter M, Bleve-Zacheo T, Davis EL, Ewbank JJ, Favery B, Grenier E, Henrissat B, Jones JT, Laudet V, Maule AG, Quesneville H, Rosso MN, Schiex T, Smant G, Weissenbach J, Wincker P. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat Biotechnol. 2008;26:909–915. - PubMed
-
- Bethony J, Brooker S, Albonico M, Geiger SM, Loukas A, Diemert D, Hotez PJ. Soil-transmitted helminth infections: ascariasis, trichuriasis, and hookworm. Lancet. 2006;367:1521–1532. - PubMed
-
- Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, Vanfleteren JR, Mackey LY, Dorris M, Frisse LM, Vida JT, Thomas WK. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–75. - PubMed
-
- Blaxter ML. Nematoda: genes, genomes and the evolution of parasitism. Adv Parasitol. 2003;54:101–195. - PubMed
-
- Blaxter ML. Nematodes (Nematoda) In: Hedges SB, Kumar S, editors. The TimeTree of Life. Oxford University Press; New York: 2009. pp. 247–250.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

