Signal-Dependent Recruitment of BRD4 to Cardiomyocyte Super-Enhancers Is Suppressed by a MicroRNA
- PMID: 27425608
- PMCID: PMC4972677
- DOI: 10.1016/j.celrep.2016.06.074
Signal-Dependent Recruitment of BRD4 to Cardiomyocyte Super-Enhancers Is Suppressed by a MicroRNA
Abstract
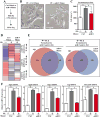
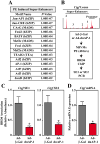
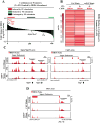
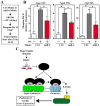
BRD4 governs pathological cardiac gene expression by binding acetylated chromatin, resulting in enhanced RNA polymerase II (Pol II) phosphorylation and transcription elongation. Here, we describe a signal-dependent mechanism for the regulation of BRD4 in cardiomyocytes. BRD4 expression is suppressed by microRNA-9 (miR-9), which targets the 3' UTR of the Brd4 transcript. In response to stress stimuli, miR-9 is downregulated, leading to derepression of BRD4 and enrichment of BRD4 at long-range super-enhancers (SEs) associated with pathological cardiac genes. A miR-9 mimic represses stimulus-dependent targeting of BRD4 to SEs and blunts Pol II phosphorylation at proximal transcription start sites, without affecting BRD4 binding to SEs that control constitutively expressed cardiac genes. These findings suggest that dynamic enrichment of BRD4 at SEs genome-wide serves a crucial role in the control of stress-induced cardiac gene expression and define a miR-dependent signaling mechanism for the regulation of chromatin state and Pol II phosphorylation.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

