The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR
- PMID: 27426517
- PMCID: PMC4980197
- DOI: 10.1016/j.cub.2016.06.002
The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR
Abstract
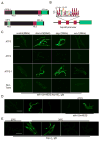
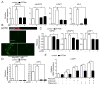
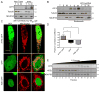
Mitochondrial dysfunction is pervasive in human pathologies such as neurodegeneration, diabetes, cancer, and pathogen infections as well as during normal aging. Cells sense and respond to mitochondrial dysfunction by activating a protective transcriptional program known as the mitochondrial unfolded protein response (UPR(mt)), which includes genes that promote mitochondrial protein homeostasis and the recovery of defective organelles [1, 2]. Work in Caenorhabditis elegans has shown that the UPR(mt) is regulated by the transcription factor ATFS-1, which is regulated by organelle partitioning. Normally, ATFS-1 accumulates within mitochondria, but during respiratory chain dysfunction, high levels of reactive oxygen species (ROS), or mitochondrial protein folding stress, a percentage of ATFS-1 accumulates in the cytosol and traffics to the nucleus where it activates the UPR(mt) [2]. While similar transcriptional responses have been described in mammals [3, 4], how the UPR(mt) is regulated remains unclear. Here, we describe a mammalian transcription factor, ATF5, which is regulated similarly to ATFS-1 and induces a similar transcriptional response. ATF5 expression can rescue UPR(mt) signaling in atfs-1-deficient worms requiring the same UPR(mt) promoter element identified in C. elegans. Furthermore, mammalian cells require ATF5 to maintain mitochondrial activity during mitochondrial stress and promote organelle recovery. Combined, these data suggest that regulation of the UPR(mt) is conserved from worms to mammals.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

