Genomic Recoding Broadly Obstructs the Propagation of Horizontally Transferred Genetic Elements
- PMID: 27426981
- PMCID: PMC5568630
- DOI: 10.1016/j.cels.2016.06.009
Genomic Recoding Broadly Obstructs the Propagation of Horizontally Transferred Genetic Elements
Abstract
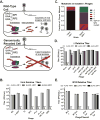
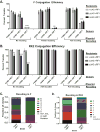
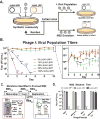
Horizontally transferred genetic elements such as viruses and conjugative plasmids move DNA between organisms, increasing genetic diversity but destabilizing engineered biological systems. Here, we used a genomically recoded Escherichia coli strain lacking UAG stop codons and the recognition protein release factor 1 to study how an alternative genetic code influences horizontally transferred genetic element propagation. The alternative genetic code conferred resistance to multiple viruses (λ, M13, P1, MS2) at titers up to 10(11) PFU/ml and impaired conjugative plasmids (F and RK2) up to 10(5)-fold. By recoding UAG codons to UAA in viruses and plasmids, we restored viral infectivity and conjugative function. Propagating viruses on a mixed community of cells with standard and alternative genetic codes reduced viral titer, and over time viruses adapted to the alternative genetic code. This work demonstrates that altering the genetic code broadly obstructs the propagation of horizontally transferred genetic elements and supports the use of genomic recoding as a strategy to stabilize engineered biological systems.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
References
-
- Baltrus DA. Exploring the costs of horizontal gene transfer. Trends Ecol Evol. 2013;28:489–495. - PubMed
-
- Bethencourt V. Virus stalls Genzyme plant. Nat Biotech. 2009;27:681–681.
-
- Calendar R. The bacteriophages. 2. Oxford ; New York: Oxford University Press; 2006.
-
- Carlson R. Estimating the biotech sector's contribution to the US economy. Nature biotechnology. 2016;34:247–255. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

