Relational Network for Knowledge Discovery through Heterogeneous Biomedical and Clinical Features
- PMID: 27427091
- PMCID: PMC4947904
- DOI: 10.1038/srep29915
Relational Network for Knowledge Discovery through Heterogeneous Biomedical and Clinical Features
Abstract
Biomedical big data, as a whole, covers numerous features, while each dataset specifically delineates part of them. "Full feature spectrum" knowledge discovery across heterogeneous data sources remains a major challenge. We developed a method called bootstrapping for unified feature association measurement (BUFAM) for pairwise association analysis, and relational dependency network (RDN) modeling for global module detection on features across breast cancer cohorts. Discovered knowledge was cross-validated using data from Wake Forest Baptist Medical Center's electronic medical records and annotated with BioCarta signaling signatures. The clinical potential of the discovered modules was exhibited by stratifying patients for drug responses. A series of discovered associations provided new insights into breast cancer, such as the effects of patient's cultural background on preferences for surgical procedure. We also discovered two groups of highly associated features, the HER2 and the ER modules, each of which described how phenotypes were associated with molecular signatures, diagnostic features, and clinical decisions. The discovered "ER module", which was dominated by cancer immunity, was used as an example for patient stratification and prediction of drug responses to tamoxifen and chemotherapy. BUFAM-derived RDN modeling demonstrated unique ability to discover clinically meaningful and actionable knowledge across highly heterogeneous biomedical big data sets.
Figures

 . Gray regions represent non-testable associations. (B) Validation of discovered associations using the EMR of Wake Forest Baptist Medical Center (WakeOne) presented in
. Gray regions represent non-testable associations. (B) Validation of discovered associations using the EMR of Wake Forest Baptist Medical Center (WakeOne) presented in  . Associations between Oncotype DX score and histologic grade, and between Oncotype DX score and progesterone status, are shown as examples.
. Associations between Oncotype DX score and histologic grade, and between Oncotype DX score and progesterone status, are shown as examples.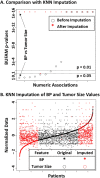


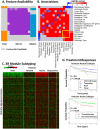
 . The yellow box highlights common associations shared by two groups of associations. (C) Patient subtyping using the BioCarta signatures (top region) associated with the ER module. These signatures revealed four subtypes: Immune Inert, Neutral, Active, and Responsive. (D) Differential treatment responses assessed by Kaplan-Meier survival analysis of the WFCCC cohort, using distant-metastasis-free survival time as the index. Survival curves of the Immune Neutral (top) and Active (bottom) Subtypes under different treatments are presented and labeled with patient numbers and log-rank test p-values. More details are given in the Supplement.
. The yellow box highlights common associations shared by two groups of associations. (C) Patient subtyping using the BioCarta signatures (top region) associated with the ER module. These signatures revealed four subtypes: Immune Inert, Neutral, Active, and Responsive. (D) Differential treatment responses assessed by Kaplan-Meier survival analysis of the WFCCC cohort, using distant-metastasis-free survival time as the index. Survival curves of the Immune Neutral (top) and Active (bottom) Subtypes under different treatments are presented and labeled with patient numbers and log-rank test p-values. More details are given in the Supplement.References
-
- Rosenthal A. et al.. Cloud computing: a new business paradigm for biomedical information sharing. Journal of Biomedical Informatics 43, 342–353 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

