Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes
- PMID: 27427974
- PMCID: PMC5079676
- DOI: 10.1080/15548627.2016.1208889
Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes
Abstract
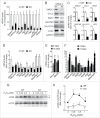
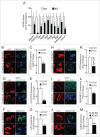
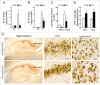
Autophagy is a highly coordinated process that is controlled at several levels including transcriptional regulation. Here, we identify the transcription factor NFE2L2/NRF2 (nuclear factor, erythroid 2 like 2) as a regulator of autophagy gene expression and its relevance in a mouse model of Alzheimer disease (AD) that reproduces impaired APP (amyloid β precursor protein) and human (Hs)MAPT/TAU processing, clearance and aggregation. We screened the chromatin immunoprecipitation database ENCODE for 2 proteins, MAFK and BACH1, that bind the NFE2L2-regulated enhancer antioxidant response element (ARE). Using a script generated from the JASPAR's consensus ARE sequence, we identified 27 putative AREs in 16 autophagy-related genes. Twelve of these sequences were validated as NFE2L2 regulated AREs in 9 autophagy genes by additional ChIP assays and quantitative RT-PCR on human and mouse cells after NFE2L2 activation with sulforaphane. Mouse embryo fibroblasts of nfe2l2-knockout mice exhibited reduced expression of autophagy genes, which was rescued by an NFE2L2 expressing lentivirus, and impaired autophagy flux when exposed to hydrogen peroxide. NFE2L2-deficient mice co-expressing HsAPPV717I and HsMAPTP301L, exhibited more intracellular aggregates of these proteins and reduced neuronal levels of SQSTM1/p62, CALCOCO2/NDP52, ULK1, ATG5 and GABARAPL1. Also, colocalization of HsAPPV717I and HsMAPTP301L with the NFE2L2-regulated autophagy marker SQSTM1/p62 was reduced in the absence of NFE2L2. In AD patients, neurons expressing high levels of APP or MAPT also expressed SQSTM1/p62 and nuclear NFE2L2, suggesting their attempt to degrade intraneuronal aggregates through autophagy. This study shows that NFE2L2 modulates autophagy gene expression and suggests a new strategy to combat proteinopathies.
Keywords: Alzheimer disease; Tau; amyloid precursor protein; neurodegenerative diseases; neuroprotection; oxidative stress; proteostasis.
Figures




References
-
- Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell 2011; 146:682-95; PMID:21884931; http://dx.doi.org/10.1016/j.cell.2011.07.030 - DOI - PubMed
-
- Ciechanover A, Kwon YT. Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Exp Mol Med 2015; 47:e147; PMID:25766616; http://dx.doi.org/10.1038/emm.2014.117 - DOI - PMC - PubMed
-
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000; 290:1717-21; PMID:11099404; http://dx.doi.org/10.1126/science.290.5497.1717 - DOI - PMC - PubMed
-
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13:132-41; PMID:21258367; http://dx.doi.org/10.1038/ncb2152 - DOI - PMC - PubMed
-
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al.. TFEB links autophagy to lysosomal biogenesis. Science 2011; 332:1429-33; PMID:21617040; http://dx.doi.org/10.1126/science.1204592 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
