p21 mediates macrophage reprogramming through regulation of p50-p50 NF-κB and IFN-β
- PMID: 27427981
- PMCID: PMC4966310
- DOI: 10.1172/JCI83404
p21 mediates macrophage reprogramming through regulation of p50-p50 NF-κB and IFN-β
Abstract
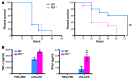
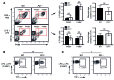
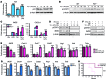
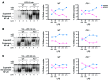
M1 and M2 macrophage phenotypes, which mediate proinflammatory and antiinflammatory functions, respectively, represent the extremes of immunoregulatory plasticity in the macrophage population. This plasticity can also result in intermediate macrophage states that support a balance between these opposing functions. In sepsis, M1 macrophages can compensate for hyperinflammation by acquiring an M2-like immunosuppressed status that increases the risk of secondary infection and death. The M1 to M2 macrophage reprogramming that develops during LPS tolerance resembles the pathological antiinflammatory response to sepsis. Here, we determined that p21 regulates macrophage reprogramming by shifting the balance between active p65-p50 and inhibitory p50-p50 NF-κB pathways. p21 deficiency reduced the DNA-binding affinity of the p50-p50 homodimer in LPS-primed and -rechallenged macrophages, impairing their ability to attenuate IFN-β production and acquire an M2-like hyporesponsive status. High p21 levels in sepsis patients correlated with low IFN-β expression, and p21 knockdown in human monocytes corroborated its role in IFN-β regulation. The data demonstrate that p21 adjusts the equilibrium between p65-p50 and p50-p50 NF-κB pathways to mediate macrophage plasticity in LPS tolerance. Identifying p21-related pathways involved in monocyte reprogramming may lead to potential targets for sepsis treatment.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

