Schlafen 4-expressing myeloid-derived suppressor cells are induced during murine gastric metaplasia
- PMID: 27427984
- PMCID: PMC4966326
- DOI: 10.1172/JCI82529
Schlafen 4-expressing myeloid-derived suppressor cells are induced during murine gastric metaplasia
Abstract
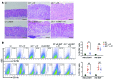
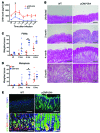
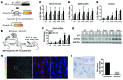
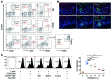
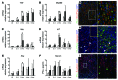
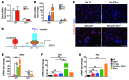
Chronic Helicobacter pylori infection triggers neoplastic transformation of the gastric mucosa in a small subset of patients, but the risk factors that induce progression to gastric metaplasia have not been identified. Prior to cancer development, the oxyntic gastric glands atrophy and are replaced by metaplastic cells in response to chronic gastritis. Previously, we identified schlafen 4 (Slfn4) as a GLI1 target gene and myeloid differentiation factor that correlates with spasmolytic polypeptide-expressing metaplasia (SPEM) in mice. Here, we tested the hypothesis that migration of SLFN4-expressing cells from the bone marrow to peripheral organs predicts preneoplastic changes in the gastric microenvironment. Lineage tracing in Helicobacter-infected Slfn4 reporter mice revealed that SLFN4+ cells migrated to the stomach, where they exhibited myeloid-derived suppressor cell (MDSC) markers and acquired the ability to inhibit T cell proliferation. SLFN4+ MDSCs were not observed in infected GLI1-deficient mice. Overexpression of sonic hedgehog ligand (SHH) in infected WT mice accelerated the appearance of SLFN4+ MDSCs in the gastric corpus. Similarly, in the stomachs of H. pylori-infected patients, the human SLFN4 ortholog SLFN12L colocalized to cells that expressed MDSC surface markers CD15+CD33+HLA-DRlo. Together, these results indicate that SLFN4 marks a GLI1-dependent population of MDSCs that predict a shift in the gastric mucosa to a metaplastic phenotype.
Figures

References
-
- Haenszel W, Correa P. Developments in the epidemiology of stomach cancer over the past decade. Cancer Res. 1975;35(11 pt 2):3452–3459. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

