Small Molecule Inhibitor of CBFβ-RUNX Binding for RUNX Transcription Factor Driven Cancers
- PMID: 27428424
- PMCID: PMC4919611
- DOI: 10.1016/j.ebiom.2016.04.032
Small Molecule Inhibitor of CBFβ-RUNX Binding for RUNX Transcription Factor Driven Cancers
Erratum in
-
Corrigendum to: "Small Molecule Inhibitor of CBFβ-RUNX Binding for RUNX Transcription Factor Driven Cancers" [EBioMedicine 8 (2016) 117-131].EBioMedicine. 2017 Nov;25:188. doi: 10.1016/j.ebiom.2017.10.029. Epub 2017 Oct 31. EBioMedicine. 2017. PMID: 29104075 Free PMC article. No abstract available.
Abstract
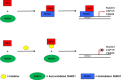
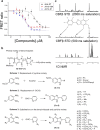
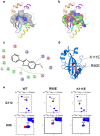
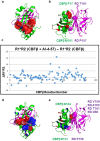
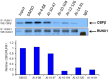
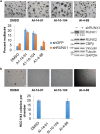
Transcription factors have traditionally been viewed with skepticism as viable drug targets, but they offer the potential for completely novel mechanisms of action that could more effectively address the stem cell like properties, such as self-renewal and chemo-resistance, that lead to the failure of traditional chemotherapy approaches. Core binding factor is a heterodimeric transcription factor comprised of one of 3 RUNX proteins (RUNX1-3) and a CBFβ binding partner. CBFβ enhances DNA binding of RUNX subunits by relieving auto-inhibition. Both RUNX1 and CBFβ are frequently mutated in human leukemia. More recently, RUNX proteins have been shown to be key players in epithelial cancers, suggesting the targeting of this pathway could have broad utility. In order to test this, we developed small molecules which bind to CBFβ and inhibit its binding to RUNX. Treatment with these inhibitors reduces binding of RUNX1 to target genes, alters the expression of RUNX1 target genes, and impacts cell survival and differentiation. These inhibitors show efficacy against leukemia cells as well as basal-like (triple-negative) breast cancer cells. These inhibitors provide effective tools to probe the utility of targeting RUNX transcription factor function in other cancers.
Keywords: CBFβ; Leukemia; PPI; RUNX; Transcription factor inhibitor; Triple negative breast cancer.
Copyright © 2016 The Authors. Published by Elsevier B.V. All rights reserved.
Figures


References
-
- Adya N., Castilla L.H., Liu P.P. Function of CBFbeta/Bro proteins. Semin. Cell Dev. Biol. 2000;11:361–368. - PubMed
-
- Arkin M.R., Whitty A. The road less traveled: modulating signal transduction enzymes by inhibiting their protein-protein interactions. Curr. Opin. Chem. Biol. 2009;13:284–290. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

