Genome-Wide Analysis of Codon Usage Bias in Epichloë festucae
- PMID: 27428961
- PMCID: PMC4964511
- DOI: 10.3390/ijms17071138
Genome-Wide Analysis of Codon Usage Bias in Epichloë festucae
Abstract
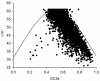
Analysis of codon usage data has both practical and theoretical applications in understanding the basics of molecular biology. Differences in codon usage patterns among genes reflect variations in local base compositional biases and the intensity of natural selection. Recently, there have been several reports related to codon usage in fungi, but little is known about codon usage bias in Epichloë endophytes. The present study aimed to assess codon usage patterns and biases in 4870 sequences from Epichloë festucae, which may be helpful in revealing the constraint factors such as mutation or selection pressure and improving the bioreactor on the cloning, expression, and characterization of some special genes. The GC content with 56.41% is higher than the AT content (43.59%) in E. festucae. The results of neutrality and effective number of codons plot analyses showed that both mutational bias and natural selection play roles in shaping codon usage in this species. We found that gene length is strongly correlated with codon usage and may contribute to the codon usage patterns observed in genes. Nucleotide composition and gene expression levels also shape codon usage bias in E. festucae. E. festucae exhibits codon usage bias based on the relative synonymous codon usage (RSCU) values of 61 sense codons, with 25 codons showing an RSCU larger than 1. In addition, we identified 27 optimal codons that end in a G or C.
Keywords: Epichloë festucae; codon usage bias; grass endophyte; natural selection; optimal codons.
Figures

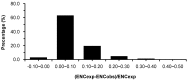
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

