Ligand and Structure-Based Approaches for the Identification of Peptide Deformylase Inhibitors as Antibacterial Drugs
- PMID: 27428963
- PMCID: PMC4964514
- DOI: 10.3390/ijms17071141
Ligand and Structure-Based Approaches for the Identification of Peptide Deformylase Inhibitors as Antibacterial Drugs
Abstract
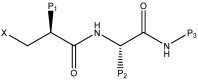
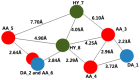
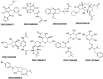
Peptide deformylase (PDF) is a metalloprotease catalyzing the removal of a formyl group from newly synthesized proteins, which makes it an important antibacterial drug target. Given the importance of PDF inhibitors like actinonin in antibacterial drug discovery, several reported potent PDF inhibitors were used to develop pharmacophore models using the Galahad module of Sybyl 7.1 software. Generated pharmacophore models were composed of two donor atom centers, four acceptor atom centers and two hydrophobic groups. Model-1 was screened against the Zinc database and several compounds were retrieved as hits. Compounds with Qfit values of more than 60 were employed to perform a molecular docking study with the receptor Escherichia coli PDF, then compounds with docking score values of more than 6 were used to predict the in silico pharmacokinetic and toxicity risk via OSIRIS property explorer. Two known PDF inhibitors were also used to perform a molecular docking study with E. coli PDF as reference molecules. The results of the molecular docking study were validated by reproducing the crystal structure of actinonin. Molecular docking and in silico pharmacokinetic and toxicity prediction studies suggested that ZINC08740166 has a relatively high docking score of 7.44 and a drug score of 0.78.
Keywords: antibacterial drug; high-throughput virtual screening; molecular docking; peptide deformylase; pharmacophore model.
Figures

References
-
- Briers Y., Walmagh M., Grymonprez B., Biebl M., Pirnay J.P., Defraine V., Michiels J., Cenens W., Aertsen A., Miller S., et al. Art-175 is a highly efficient antibacterial against multidrug-resistant strains and persisters of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2014;58:3774–3784. doi: 10.1128/AAC.02668-14. - DOI - PMC - PubMed
-
- Osborne C.S., Neckermann G., Fischer E., Pecanka R., Yu D., Manni K., Goldovitz J., Amaral K., Dzink-Fox J., Ryder N.S. In vivo characterization of the peptide deformylase inhibitor LBM415 in murine infection models. Antimicrob. Agents Chemother. 2009;53:3777–3781. doi: 10.1128/AAC.00026-09. - DOI - PMC - PubMed
-
- Haeili M., Moore C., Davis C.J., Cochran J.B., Shah S., Shrestha T.B., Zhang Y., Bossmann S.H., Benjamin W.H., Kutsch O., et al. Copper complexation screen reveals compounds with potent antibiotic properties against methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2014;58:3727–3736. doi: 10.1128/AAC.02316-13. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

