Tunable regulation of CREB DNA binding activity couples genotoxic stress response and metabolism
- PMID: 27431323
- PMCID: PMC5175338
- DOI: 10.1093/nar/gkw643
Tunable regulation of CREB DNA binding activity couples genotoxic stress response and metabolism
Abstract
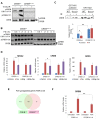
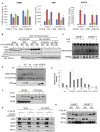
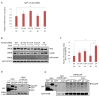
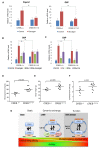
cAMP response element binding protein (CREB) is a key regulator of glucose metabolism and synaptic plasticity that is canonically regulated through recruitment of transcriptional coactivators. Here we show that phosphorylation of CREB on a conserved cluster of Ser residues (the ATM/CK cluster) by the DNA damage-activated protein kinase ataxia-telangiectasia-mutated (ATM) and casein kinase1 (CK1) and casein kinase2 (CK2) positively and negatively regulates CREB-mediated transcription in a signal dependent manner. In response to genotoxic stress, phosphorylation of the ATM/CK cluster inhibited CREB-mediated gene expression, DNA binding activity and chromatin occupancy proportional to the number of modified Ser residues. Paradoxically, substoichiometric, ATM-independent, phosphorylation of the ATM/CK cluster potentiated bursts in CREB-mediated transcription by promoting recruitment of the CREB coactivator, cAMP-regulated transcriptional coactivators (CRTC2). Livers from mice expressing a non-phosphorylatable CREB allele failed to attenuate gluconeogenic genes in response to DNA damage or fully activate the same genes in response to glucagon. We propose that phosphorylation-dependent regulation of DNA binding activity evolved as a tunable mechanism to control CREB transcriptional output and promote metabolic homeostasis in response to rapidly changing environmental conditions.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Coregulated ataxia telangiectasia-mutated and casein kinase sites modulate cAMP-response element-binding protein-coactivator interactions in response to DNA damage.J Biol Chem. 2007 Mar 2;282(9):6283-91. doi: 10.1074/jbc.M610674200. Epub 2007 Jan 5. J Biol Chem. 2007. PMID: 17209043
-
Conserved and distinct modes of CREB/ATF transcription factor regulation by PP2A/B56gamma and genotoxic stress.PLoS One. 2010 Aug 13;5(8):e12173. doi: 10.1371/journal.pone.0012173. PLoS One. 2010. PMID: 20730097 Free PMC article.
-
Cyclin-dependent kinase 1-dependent phosphorylation of cAMP response element-binding protein decreases chromatin occupancy.J Biol Chem. 2013 Aug 16;288(33):23765-75. doi: 10.1074/jbc.M113.464057. Epub 2013 Jun 27. J Biol Chem. 2013. PMID: 23814058 Free PMC article.
-
Transcription factors and coactivators controlling nutrient and hormonal regulation of hepatic gluconeogenesis.Int J Biochem Cell Biol. 2012 Jan;44(1):33-45. doi: 10.1016/j.biocel.2011.10.001. Epub 2011 Oct 8. Int J Biochem Cell Biol. 2012. PMID: 22004992 Review.
-
Regulation of somatostatin gene transcription by cyclic adenosine monophosphate.Metabolism. 1996 Aug;45(8 Suppl 1):4-7. doi: 10.1016/s0026-0495(96)90068-2. Metabolism. 1996. PMID: 8769368 Review.
Cited by
-
Fused in sarcoma regulates DNA replication timing and kinetics.J Biol Chem. 2021 Sep;297(3):101049. doi: 10.1016/j.jbc.2021.101049. Epub 2021 Aug 8. J Biol Chem. 2021. PMID: 34375640 Free PMC article.
-
Linking CREB function with altered metabolism in murine fibroblast-based model cell lines.Oncotarget. 2017 Oct 27;8(57):97439-97463. doi: 10.18632/oncotarget.22135. eCollection 2017 Nov 14. Oncotarget. 2017. PMID: 29228623 Free PMC article.
-
Glutamine-rich regions of the disordered CREB transactivation domain mediate dynamic intra- and intermolecular interactions.Proc Natl Acad Sci U S A. 2023 Nov 21;120(47):e2313835120. doi: 10.1073/pnas.2313835120. Epub 2023 Nov 14. Proc Natl Acad Sci U S A. 2023. PMID: 37971402 Free PMC article.
-
Increased mAb production in amplified CHO cell lines is associated with increased interaction of CREB1 with transgene promoter.Curr Res Biotechnol. 2019 Nov;1:49-57. doi: 10.1016/j.crbiot.2019.09.001. Epub 2019 Oct 5. Curr Res Biotechnol. 2019. PMID: 32577618 Free PMC article.
-
ATM-dependent pathways of chromatin remodelling and oxidative DNA damage responses.Philos Trans R Soc Lond B Biol Sci. 2017 Oct 5;372(1731):20160283. doi: 10.1098/rstb.2016.0283. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28847820 Free PMC article. Review.
References
-
- Impey S., McCorkle S.R., Cha-Molstad H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. - PubMed
-
- Zhang X., Odom D.T., Koo S.-H., Conkright M.D., Canettieri G., Best J., Chen H., Jenner R., Herbolsheimer E., Jacobsen E., et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4459–4464. - PMC - PubMed
-
- Oh K.-J., Han H.-S., Kim M.-J., Koo S.-H. Transcriptional regulators of hepatic gluconeogenesis. Arch. Pharm. Res. 2013;36:189–200. - PubMed
-
- Kida S., Serita T. Functional roles of CREB as a positive regulator in the formation and enhancement of memory. Brain Res. Bull. 2014;105:17–24. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

