Tunable regulation of CREB DNA binding activity couples genotoxic stress response and metabolism
- PMID: 27431323
- PMCID: PMC5175338
- DOI: 10.1093/nar/gkw643
Tunable regulation of CREB DNA binding activity couples genotoxic stress response and metabolism
Abstract
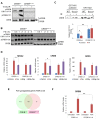
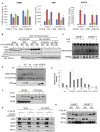
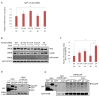
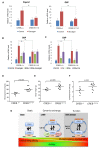
cAMP response element binding protein (CREB) is a key regulator of glucose metabolism and synaptic plasticity that is canonically regulated through recruitment of transcriptional coactivators. Here we show that phosphorylation of CREB on a conserved cluster of Ser residues (the ATM/CK cluster) by the DNA damage-activated protein kinase ataxia-telangiectasia-mutated (ATM) and casein kinase1 (CK1) and casein kinase2 (CK2) positively and negatively regulates CREB-mediated transcription in a signal dependent manner. In response to genotoxic stress, phosphorylation of the ATM/CK cluster inhibited CREB-mediated gene expression, DNA binding activity and chromatin occupancy proportional to the number of modified Ser residues. Paradoxically, substoichiometric, ATM-independent, phosphorylation of the ATM/CK cluster potentiated bursts in CREB-mediated transcription by promoting recruitment of the CREB coactivator, cAMP-regulated transcriptional coactivators (CRTC2). Livers from mice expressing a non-phosphorylatable CREB allele failed to attenuate gluconeogenic genes in response to DNA damage or fully activate the same genes in response to glucagon. We propose that phosphorylation-dependent regulation of DNA binding activity evolved as a tunable mechanism to control CREB transcriptional output and promote metabolic homeostasis in response to rapidly changing environmental conditions.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Impey S., McCorkle S.R., Cha-Molstad H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. - PubMed
-
- Zhang X., Odom D.T., Koo S.-H., Conkright M.D., Canettieri G., Best J., Chen H., Jenner R., Herbolsheimer E., Jacobsen E., et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4459–4464. - PMC - PubMed
-
- Oh K.-J., Han H.-S., Kim M.-J., Koo S.-H. Transcriptional regulators of hepatic gluconeogenesis. Arch. Pharm. Res. 2013;36:189–200. - PubMed
-
- Kida S., Serita T. Functional roles of CREB as a positive regulator in the formation and enhancement of memory. Brain Res. Bull. 2014;105:17–24. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

