Metagenomic Analysis Revealed Methylamine and Ureide Utilization of Soybean-Associated Methylobacterium
- PMID: 27431374
- PMCID: PMC5017803
- DOI: 10.1264/jsme2.ME16035
Metagenomic Analysis Revealed Methylamine and Ureide Utilization of Soybean-Associated Methylobacterium
Abstract
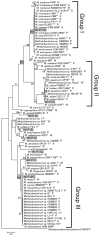
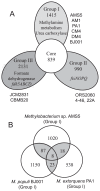
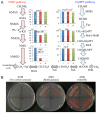
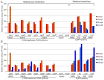
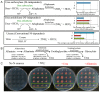
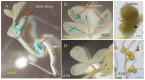
Methylobacterium inhabits the phyllosphere of a large number of plants. We herein report the results of comparative metagenome analyses on methylobacterial communities of soybean plants grown in an experimental field in Tohoku University (Kashimadai, Miyagi, Japan). Methylobacterium was identified as the most dominant genus (33%) among bacteria inhabiting soybean stems. We classified plant-derived Methylobacterium species into Groups I, II, and III based on 16S rRNA gene sequences, and found that Group I members (phylogenetically close to M. extorquens) were dominant in soybean-associated Methylobacterium. By comparing 29 genomes, we found that all Group I members possessed a complete set of genes for the N-methylglutamate pathway for methylamine utilization, and genes for urea degradation (urea carboxylase, urea amidolyase, and conventional urease). Only Group I members and soybean methylobacterial isolates grew in a culture supplemented with methylamine as the sole carbon source. They utilized urea or allantoin (a urea-related compound in legumes) as the sole nitrogen source; however, group III also utilized these compounds. The utilization of allantoin may be crucial in soybean-bacterial interactions because allantoin is a transported form of fixed nitrogen in legume plants. Soybean-derived Group I strain AMS5 colonized the model legume Lotus japonicus well. A comparison among the 29 genomes of plant-derived and other strains suggested that several candidate genes are involved in plant colonization such as csgG (curli fimbriae). Genes for the N-methylglutamate pathway and curli fimbriae were more abundant in soybean microbiomes than in rice microbiomes in the field. Based on these results, we discuss the lifestyle of Methylobacterium in the legume phyllosphere.
Figures
Similar articles
-
Methylobacterium nodulans sp. nov., for a group of aerobic, facultatively methylotrophic, legume root-nodule-forming and nitrogen-fixing bacteria.Int J Syst Evol Microbiol. 2004 Nov;54(Pt 6):2269-2273. doi: 10.1099/ijs.0.02902-0. Int J Syst Evol Microbiol. 2004. PMID: 15545469
-
Isolation and genetic characterization of Aurantimonas and Methylobacterium strains from stems of hypernodulated soybeans.Microbes Environ. 2011;26(2):172-80. doi: 10.1264/jsme2.me10203. Epub 2011 Apr 21. Microbes Environ. 2011. PMID: 21512309
-
Characterization of Methylobacterium strains isolated from the phyllosphere and description of Methylobacterium longum sp. nov.Antonie Van Leeuwenhoek. 2012 Jan;101(1):169-83. doi: 10.1007/s10482-011-9650-6. Epub 2011 Oct 11. Antonie Van Leeuwenhoek. 2012. PMID: 21986935
-
Ureide biosynthesis in legume nodules.Front Biosci. 2004 May 1;9:1374-81. doi: 10.2741/1345. Front Biosci. 2004. PMID: 14977553 Review.
-
Biotechnological and agronomic potential of endophytic pink-pigmented methylotrophic Methylobacterium spp.Biomed Res Int. 2015;2015:909016. doi: 10.1155/2015/909016. Epub 2015 Mar 10. Biomed Res Int. 2015. PMID: 25861650 Free PMC article. Review.
Cited by
-
Community Analysis-based Screening of Plant Growth-promoting Bacteria for Sugar Beet.Microbes Environ. 2021;36(2):ME20137. doi: 10.1264/jsme2.ME20137. Microbes Environ. 2021. PMID: 33907063 Free PMC article.
-
Growth Stage-dependent Bacterial Communities in Soybean Plant Tissues: Methylorubrum Transiently Dominated in the Flowering Stage of the Soybean Shoot.Microbes Environ. 2019 Dec 27;34(4):446-450. doi: 10.1264/jsme2.ME19067. Epub 2019 Aug 14. Microbes Environ. 2019. PMID: 31413227 Free PMC article.
-
Nitrogen Deficiency-induced Bacterial Community Shifts in Soybean Roots.Microbes Environ. 2021;36(3):ME21004. doi: 10.1264/jsme2.ME21004. Microbes Environ. 2021. PMID: 34234044 Free PMC article.
-
Rhizosphere Metagenomics of Paspalum scrobiculatum L. (Kodo Millet) Reveals Rhizobiome Multifunctionalities.Microorganisms. 2019 Nov 23;7(12):608. doi: 10.3390/microorganisms7120608. Microorganisms. 2019. PMID: 31771141 Free PMC article.
-
Plant-Associated Microbes: From Rhizobia To Plant Microbiomes.Microbes Environ. 2018;33(1):1-3. doi: 10.1264/jsme2.ME3301rh. Microbes Environ. 2018. PMID: 29593170 Free PMC article. No abstract available.
References
-
- Aken B.V., Peres C.M., Doty S.L., Yoon J.M., Schnoor J.L. Methylobacterium populi sp. nov., a novel aerobic, pink-pigmented, facultatively methylotrophic, methane-utilizing bacterium isolated from poplar trees (Populus deltoides×nigra DN34) Int J Syst Evol Microbiol. 2004;54:1191–1196. - PubMed
-
- Anda M., Ikeda S., Eda S., Okubo T., Sato S., Tabata S., Mitsui H., Minamisawa K. Isolation and genetic characterization of Aurantimonas and Methylobacterium strains from stems of hypernodulated soybeans. Microbes Environ. 2011;26:172–180. - PubMed
-
- Ardley J.K., O’Hara G.W., Reeve W.G., Yates R.J., Dilworth M.J., Tiwari R.P., Howieson J.G. Root nodule bacteria isolated from South African Lotononis bainesii, L. listii and L. solitudinis are species of Methylobacterium that are unable to utilize methanol. Arch Microbiol. 2009;191:311–318. - PubMed
-
- Bagni N., Tassoni A. Biosynthesis, oxidation and conjugation of aliphatic polyamines in higher plants. Amino Acids. 2001;20:301–317. - PubMed
-
- Beg B.K., Kapoor M., Mahajan L., Hoondal G.S. Microbial xylanases and their industrial applications: a review. Appl Microbiol Biotechnol. 2001;56:326–338. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

