Characterization of Enzymes Catalyzing Transformations of Cysteine S-Conjugated Intermediates in the Lincosamide Biosynthetic Pathway
- PMID: 27431934
- PMCID: PMC5253346
- DOI: 10.1002/cbic.201600223
Characterization of Enzymes Catalyzing Transformations of Cysteine S-Conjugated Intermediates in the Lincosamide Biosynthetic Pathway
Abstract
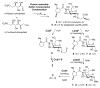
Lincosamides such as lincomycin A, celesticetin, and Bu-2545, constitute an important group of antibiotics. These natural products are characterized by a thiooctose linked to a l-proline residue, but they differ with regards to modifications of the thioacetal moiety, the pyrrolidine ring, and the octose core. Here we report that the pyridoxal 5'-phosphate-dependent enzyme CcbF (celesticetin biosynthetic pathway) is a decarboxylating deaminase that converts a cysteine S-conjugated intermediate into an aldehyde. In contrast, the homologous enzyme LmbF (lincomycin biosynthetic pathway) catalyzes C-S bond cleavage of the same intermediate to afford a thioglycoside. We show that Ccb4 and LmbG (downstream methyltransferases) convert the aldehyde and thiol intermediates into a variety of methylated lincosamide compounds including Bu-2545. The substrates used in these studies are the β-anomers of the natural substrates. The findings not only provide insight into how the biosynthetic pathway of lincosamide antibiotics can bifurcate to generate different lincosamides, but also reveal the promiscuity of the enzymes involved.
Keywords: biosynthesis; catalytic mechanisms; enzymes; lincosamides; pyridoxal 5′-phosphate.
© 2016 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures






Similar articles
-
Molecular basis for the diversification of lincosamide biosynthesis by pyridoxal phosphate-dependent enzymes.Nat Chem. 2025 Feb;17(2):256-264. doi: 10.1038/s41557-024-01687-7. Epub 2024 Dec 6. Nat Chem. 2025. PMID: 39643667 Free PMC article.
-
Studies of lincosamide formation complete the biosynthetic pathway for lincomycin A.Proc Natl Acad Sci U S A. 2020 Oct 6;117(40):24794-24801. doi: 10.1073/pnas.2009306117. Epub 2020 Sep 21. Proc Natl Acad Sci U S A. 2020. PMID: 32958639 Free PMC article.
-
Differences in PLP-Dependent Cysteinyl Processing Lead to Diverse S-Functionalization of Lincosamide Antibiotics.J Am Chem Soc. 2016 May 25;138(20):6348-51. doi: 10.1021/jacs.6b01751. Epub 2016 May 16. J Am Chem Soc. 2016. PMID: 27171737
-
Biosynthesis of Lincosamide Antibiotics: Reactions Associated with Degradation and Detoxification Pathways Play a Constructive Role.Acc Chem Res. 2018 Jun 19;51(6):1496-1506. doi: 10.1021/acs.accounts.8b00135. Epub 2018 May 24. Acc Chem Res. 2018. PMID: 29792672 Review.
-
Biosynthesis and incorporation of an alkylproline-derivative (APD) precursor into complex natural products.Nat Prod Rep. 2018 Mar 1;35(3):257-289. doi: 10.1039/c7np00047b. Epub 2018 Mar 8. Nat Prod Rep. 2018. PMID: 29517100 Review.
Cited by
-
Evolution-guided adaptation of an adenylation domain substrate specificity to an unusual amino acid.PLoS One. 2017 Dec 14;12(12):e0189684. doi: 10.1371/journal.pone.0189684. eCollection 2017. PLoS One. 2017. PMID: 29240815 Free PMC article.
-
Comparative transcriptomic analysis reveals the significant pleiotropic regulatory effects of LmbU on lincomycin biosynthesis.Microb Cell Fact. 2020 Feb 12;19(1):30. doi: 10.1186/s12934-020-01298-0. Microb Cell Fact. 2020. PMID: 32050973 Free PMC article.
-
Sulfur-Containing Metabolites from Marine and Terrestrial Fungal Sources: Origin, Structures, and Bioactivities.Mar Drugs. 2022 Dec 7;20(12):765. doi: 10.3390/md20120765. Mar Drugs. 2022. PMID: 36547912 Free PMC article. Review.
-
Molecular basis for the diversification of lincosamide biosynthesis by pyridoxal phosphate-dependent enzymes.Nat Chem. 2025 Feb;17(2):256-264. doi: 10.1038/s41557-024-01687-7. Epub 2024 Dec 6. Nat Chem. 2025. PMID: 39643667 Free PMC article.
-
Studies of lincosamide formation complete the biosynthetic pathway for lincomycin A.Proc Natl Acad Sci U S A. 2020 Oct 6;117(40):24794-24801. doi: 10.1073/pnas.2009306117. Epub 2020 Sep 21. Proc Natl Acad Sci U S A. 2020. PMID: 32958639 Free PMC article.
References
-
- Lewis C, Clapp HW, Grady JE. Antimicrob Agents Chemother. 1963:570–582.
- Mason DJ, Lewis C. Antimicrob Agents Chemother. 1964:7–12. - PubMed
- Mason DJ, Dietz A, DeBoer C. Antimicrob Agents Chemother. 1962:554–559.
- Herr RR, Bergy ME. Antimicrob Agents Chemother. 1962:560–564.
- Hoeksema H, Bannister B, Birkenmeyer RD, Kagan F, Magerlein BJ, MacKellar FA, Schroeder W, Slomp G, Herr RR. J Am Chem Soc. 1964;86:4223–4224.
- Reusser F. Antimicrob Agents Chemother. 1975;7:32–37. - PMC - PubMed
-
- Hoeksema H, Crum GF, DeVries WH. Antibiot Annu. 1955;2:837–841.
- Hoeksema H. J Am Chem Soc. 1968;90:755–757. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

